Abstract
Most patients with colorectal cancer die as a result of the disease spreading to other organs. However, no prevalent mutations have been associated with metastatic colorectal cancers1,2. Instead, particular features of the tumour microenvironment, such as lack of T-cell infiltration3, low type 1 T-helper cell (TH1) activity and reduced immune cytotoxicity2 or increased TGFβ levels4 predict adverse outcomes in patients with colorectal cancer. Here we analyse the interplay between genetic alterations and the tumour microenvironment by crossing mice bearing conditional alleles of four main colorectal cancer mutations in intestinal stem cells. Quadruple-mutant mice developed metastatic intestinal tumours that display key hallmarks of human microsatellite-stable colorectal cancers, including low mutational burden5, T-cell exclusion3 and TGFβ-activated stroma4,6,7. Inhibition of the PD-1–PD-L1 immune checkpoint provoked a limited response in this model system. By contrast, inhibition of TGFβ unleashed a potent and enduring cytotoxic T-cell response against tumour cells that prevented metastasis. In mice with progressive liver metastatic disease, blockade of TGFβ signalling rendered tumours susceptible to anti-PD-1–PD-L1 therapy. Our data show that increased TGFβ in the tumour microenvironment represents a primary mechanism of immune evasion that promotes T-cell exclusion and blocks acquisition of the TH1-effector phenotype. Immunotherapies directed against TGFβ signalling may therefore have broad applications in treating patients with advanced colorectal cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Change history
15 February 2018
An incorrect present address was listed for Adrià Cañellas; this has now been corrected online.
References
Jones, S. et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl Acad. Sci. USA 105, 4283–4288 (2008)
Mlecnik, B. et al. The tumor microenvironment and immunoscore are critical determinants of dissemination to distant metastasis. Sci. Transl. Med. 8, 327ra26 (2016)
Galon, J. et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964 (2006)
Calon, A. et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 47, 320–329 (2015)
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012)
Calon, A. et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell 22, 571–584 (2012)
Guinney, J. et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356 (2015)
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990)
Fumagalli, A. et al. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc. Natl Acad. Sci. USA 114, E2357–E2364 (2017)
O’Rourke, K. P. et al. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat. Biotechnol. 35, 577–582 (2017)
de Sousa e Melo, F. et al. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 543, 676–680 (2017)
Colnot, S. et al. Liver-targeted disruption of Apc in mice activates β-catenin signaling and leads to hepatocellular carcinomas. Proc. Natl Acad. Sci. USA 101, 17216–17221 (2004)
Jackson, E. L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001)
Marino, S., Vooijs, M., van Der Gulden, H., Jonkers, J. & Berns, A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14, 994–1004 (2000)
Levéen, P. et al. Induced disruption of the transforming growth factor beta type II receptor gene in mice causes a lethal inflammatory disorder that is transplantable. Blood 100, 560–568 (2002)
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007)
Isella, C. et al. Stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 47, 312–319 (2015)
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015)
Fujii, M. et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell 18, 827–838 (2016)
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013)
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013)
Rodón, J. et al. First-in-human dose study of the novel transforming growth factor-β receptor I kinase inhibitor LY2157299 monohydrate in patients with advanced cancer and glioma. Clin. Cancer Res. 21, 553–560 (2015)
Llosa, N. J. et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 5, 43–51 (2015)
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015)
Gorelik, L. & Flavell, R. A. Abrogation of TGFβ signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity 12, 171–181 (2000)
Marie, J. C., Liggitt, D. & Rudensky, A. Y. Cellular mechanisms of fatal early-onset autoimmunity in mice with the T cell-specific targeting of transforming growth factor-β receptor. Immunity 25, 441–454 (2006)
Thomas, D. A. & Massagué, J. TGF-β directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 8, 369–380 (2005)
Mehal, W. Z., Sheikh, S. Z., Gorelik, L. & Flavell, R. A. TGF-β signaling regulates CD8+ T cell responses to high- and low-affinity TCR interactions. Int. Immunol. 17, 531–538 (2005)
Chen, D. S. & Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330 (2017)
Joyce, J. A. & Fearon, D. T. T cell exclusion, immune privilege, and the tumor microenvironment. Science 348, 74–80 (2015)
Ruzankina, Y. et al. Deletion of the developmentally essential gene Atr in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1, 113–126 (2007)
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007)
R Core Team. R: a Language and Environment for Statistical Computing http://www.r-project.org (R Foundation for Statistical Computing, 2017)
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2009)
Warren, R. S., Yuan, H., Matli, M. R., Gillett, N. A. & Ferrara, N. Regulation by vascular endothelial growth factor of human colon cancer tumorigenesis in a mouse model of experimental liver metastasis. J. Clin. Invest. 95, 1789–1797 (1995)
Céspedes, M. V. et al. Orthotopic microinjection of human colon cancer cells in nude mice induces tumor foci in all clinically relevant metastatic sites. Am. J. Pathol. 170, 1077–1085 (2007)
Hothorn, T., Hornik, K., Van der Wiel, M. A. & Zeileis, A. A Lego system for conditional inference. Am. Stat. 60, 257–263 (2006)
Gelman, A. & Hill, J. Data analysis using regression and multilevel/hierarchical models. J. Stat. Softw. 30, 1–5 (2009)
Bates, D., Maechler, M., Bolker, B. M. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015)
Kuznetsova, A., Brockhoff, P. B. & Christensen, R. H. B. lmerTest Package: tests in linear mixed effects models J. Stat. Softw. 82, 1–26 (2015)
Abbas, A. R. et al. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 6, 319–331 (2005)
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995)
Gentleman, R., Carey, V., Huber, W., Irizarry, R. & Dudoit, S. (eds) Bioinformatics and Computational Biology Solutions using R and Bioconductor. https://doi.org/10.1007/0-387-29362-0 (Springer, 2005)
Acknowledgements
We thank E. Sancho for critical reading of this manuscript, all members of the Batlle laboratory for support and discussions, and members of the López-Bigas laboratory for help with revising the manuscript. We are grateful for the assistance of the IRB Barcelona core facilities for histopathology, functional genomics, mouse mutant and advanced digital microscopy; the flow cytometry and animal facilities of the UB/PCB; and the CRG genomic unit. D.V.F.T. held a Juan de la Cierva postdoctoral fellowship from MINECO. This work was supported by grants from the Doctor Josef Steiner Foundation, ERC advanced grant 340176, Instituto de Salud Carlos III, Olga Torres Foundation, BBVA Foundation, grant SAF-2014-53784 (MINECO) and by Fundación Botín. IRB Barcelona is the recipient of a Severo Ochoa Award of Excellence from MINECO.
Author information
Authors and Affiliations
Contributions
D.V.F.T., S.P.-P., D.S. and A.Cal. performed animal husbandry and genotyping; M.S. performed immunohistochemistry; D.V.F.T. and M.I. analysed histopathology. D.V.F.T. generated MTOs, which D.V.F.T. and D.S. characterized in vitro; D.S. performed CRISPR experiments; C.S.-O.A. and A.B.-L. performed exome and RNA-seq analyses, other bioinformatics (CMS classifier and patient data), and statistical analyses. S.P.-P., D.V.F.T., J.B.-R., A.Cañ. and X.H.-M. performed mouse isografting; D.V.F.T., D.S. and J.B.-R. quantified immunohistochemistry. D.B., J.A.M. and A.R. synthesized galunisertib. D.V.F.T. coordinated and performed animal treatments and analysed the data. D.V.F.T., J.B.-R., S.I., E.I.R. and A.R.N. performed immunophenotyping experiments. E.B. and D.V.F.T. conceived the study, coordinated experiments and wrote the manuscript. E.B. supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks L. Vermeulen and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Additional description of the genetic mouse models.
a, Kaplan–Meier survival curves for LA (n = 18 mice), LAK (n = 10), LAKT (n = 11) and LAKTP (n = 16) mice, as days post-tamoxifen induction of recombination in intestinal stem cells. Median survival is indicated in parentheses; Mantel–Cox test. b, Number of tumours (adenomas and carcinomas) in the large intestine, by genotype. Numbers for individual mice (n = 9 (LA), 8 (LAK, LAT and LAKT), 7 (LAP and LAKP), 10 (LATP), and 13 (LAKTP)) are indicated, as well as mean ± s.e.m. c, Number and grade of adenocarcinomas per individual mouse (bars), by genotype. P value for pairwise comparisons of tumour number: LA versus 2×, 0.0198; LA versus 3×, 0.0337; LA versus LAKTP, 0.0001; 2× versus LAKTP, 0.0051; and 3× versus LAKTP, 0.0014; n = 9 mice (LA), 8 (LAK, LAT, LAKT), 7 (LAP, LAKP), 10 (LATP), 13 (LAKTP); two-sided Mann–Whitney U test. d–i, Example images of tumour types scored by the TNM criteria, representative of 23 independent experiments. Arrows indicate invasive cells or glands, absent in an adenoma (d), restricted to the mucosa in an in situ tumour (Tis, e), breaching the muscularis mucosae (MM) in a submucosal invasive tumour (T1, f), penetrating the muscle layer (ML) in an intramuscular invasive tumour (T2, g), reaching the serosa layer beyond the ML in a submuscular invasive tumour (T3, h), or infringing all layers including the serosa in a subserosal or T4 tumour (i). j, Representative micrograph of an haematoxylin- and eosin-stained invasive subserosal adenocarcinoma (T4) of an LAKTP mouse. Carcinoma (CA) glands are indicated, invading through the muscle layer (ML). k–n, Examples of spontaneous metastases (Met) in the mesentery, diaphragm, liver and lung of induced LAKTP mice, each image is representative of two experiments. Scale bars, 500 μm.
Extended Data Figure 2 Analysis of the TME of LAKTP adenocarcinomas in the genetic model.
a, Immune infiltration in LAKTP carcinomas. Immunohistochemistry for CD3, indicating infiltrating T cells, in an intestinal adenocarcinoma of an LAKTP mouse. Insets show a magnified view of normal mucosa and tumour centre (TC). Arrowheads indicate T cells. b–d, Markers of TGFβ activation and poor prognosis in LAKTP carcinomas in a representative section of intestine with two aggressive invasive adenocarcinomas. b, Staining for pSMAD3. Note nuclear staining in stromal cells of the tumour centre. c, Staining for CALD1. Note that it is expressed in the muscle layer throughout the intestine, and in a subset of cells in the stroma of the normal mucosa. In the tumour centre of invasive carcinomas, the staining intensity is clearly higher, especially in fibroblast-like cells. d, Staining for IGFBP7, which is barely detectable in the normal mucosa. Expression is strong in the TME of the tumour centre. Images are representative of four independent experiments Scale bars, 1 mm (b–d, top); 500 μm (a, left; b–d, middle); 100 μm (all other panels).
Extended Data Figure 3 Niche factor analysis of MTOs in vitro.
Organoid formation assay in different media with representative MTOs from LA and LAKTP mice. a, Images obtained by automated plate scanning, z-stack projection and stitching of the niche factor requirement assay. Shown are full medium, medium without EGF and medium without galunisertib (Gal) but with TGFβ1. b, Results from automated organoid detection for the MTOs shown in a, each data point shows the number of organoids in three replicate wells for representative analyses. Data are shown as mean ± s.e.m. c, Organoid diameter was calculated from detected pixel area. Data points are individual organoids, pooled from triplicate wells. Violin plots and medians are shown. d, Summary of all data obtained from different LAKTP MTOs analysed in triplicates, from both primary tumours and liver metastases. Data (individual values and Tukey box plots) are normalized to the mean of full medium. Scale bars, 1 mm.
Extended Data Figure 4 Histology and TME of primary tumours from orthotopically isografted LAKTP MTOs.
a, b, Haematoxylin and eosin staining of a LAKTP-T4 carcinoma in the genetic model (a) and of a recapitulated adenocarcinoma developed from orthotopically isografted LAKTP MTO93 (b). Liver and lymph node (LN) metastasis observed in orthotopic isografted mouse (b, right panels). Iso, primary isograft. c–f, Markers of poor prognosis in isografted primary tumours. c, Immunohistochemistry for the T-cell marker CD3 (c, main panel and right, bottom right two panels) and haematoxylin and eosin stain (c, bottom left) of an orthotopic isograft, with typical glandular differentiation. Note that there are few infiltrating T cells in the tumour centre (c, bottom right), compared to the periphery and the normal mucosa (c, middle right). d–f, An orthotopically isografted, invasive primary tumour, stained for pSMAD2 (d, arrows indicate positive stromal cells), CALD1 (e) and IGFBP7 (f). Images represent three independent experiments. Scale bars, 1 mm (c, main and bottom left); 500 μm (all other panels).
Extended Data Figure 5 Schematic of the CSM4 classifier, using the k-TSP algorithm.
a, A metacohort of four pooled human CRC datasets with CMS annotation is used to find upregulated and downregulated genes in the CMS4 subtype (434 with >1.5-fold change and 300 with <0.8-fold change). To ensure usability in mouse samples, candidate genes are filtered by high-confidence homology between the two species. The k-TSP algorithm selects from among the up- and down-regulated genes the optimal set of pairs that correctly classifies CMS4 samples. The resulting classifier is applied to mouse samples by comparing the expression of pairs of genes. For each sample, the score is increased by one unit if the expression of the ‘Up’ gene is greater than that of the ‘Down’ gene. The maximum score for CMS4 classification is 14. b, Cross-validation by a leave-one-out approach. To assess performance, we repeatedly trained a classifier on three out of the four human datasets and tested using the remaining dataset. c, Results of the leave-one-out analysis. Area under the curve (AUC) was computed after re-scaling the classifier scores to the range between zero and one. Right, receiver operating characteristic curve.
Extended Data Figure 6 Genetic and functional analyses of MTOs.
a, Whole-exome sequencing of MTOs. Number of reads for the recombined exons in reads per kilobase per million mapped reads (RPKM) in MTOs and their matched host tail DNA (Con). Mouse genotypes are indicated. b, Overview of the different LAKTP MTOs analysed, including experimental metastatic descendance (indicated by arrows), and heat map and table of the number of indicated type of somatic mutation per Mb of sequenced exome. c, Mutational signatures21 in the MTOs and mouse monolayer cell lines CT26 and MC38. As in human MSS CRC, mutations in MTOs (but not in the cell lines) are predominantly of type 1. d, mRNA expression of intestinal epithelial genes Cdx1, Cdx2 and Krt20 in cultured MTOs derived from primary tumours MTO34, 68 and 93, and spontaneous liver metatasis MTO94, compared to mouse liver fibroblasts. Values are relative to 2-DCt for Ppia, normalized to the value for MTO93; and are shown as three technical replicates with mean ± s.e.m. e, Number of liver nodules after intrasplenic injection of primary LAKTP tumour and spontaneous or experimental liver metastasis (LiM)-derived MTOs in syngeneic C57BL/6J mice. Data points represent individual mice; data are mean ± s.e.m. P values (95% confidence interval) are MTO34 (n = 10 mice) versus MTO48 (n = 9): 0.0325 (−2−0), MTO48 versus MTO138 (n = 10): 0.0305 (−108–0); MTO34 versus MTO138: 0.0031 (−108−0); MTO68 (n = 11) versus MTO129 (n = 8): 3.2 × 10−4 (−207 to −30); MTO93 (n = 10) versus MTO94 (n = 10): 1.1 × 10−4 (−60 to −19); MTO93 versus MTO140 (n = 9): 2.2 × 10−5 (−149 to −81); MTO93 versus MTO163 (n = 8): 9.1 × 10−5 (−220 to −31); two-sided Mann–Whitney U test. f, Number of liver nodules (mean ± s.e.m.) after inoculation of disaggregated MTO129 or MTO138 directly into the portal vein in syngeneic animals. Data are from individual mice, n = 6 per condition. 95% confidence interval: MTO129, (−223−0) and MTO138 (−805 to −66); two-sided Mann–Whitney U test. g, Number of liver nodules (mean ± s.e.m.) of intrasplenically injected 3× and 4× MTOs, n = 22 mice (MTO54), 20 (MTO220), 5 (MTO221), 7 (MTO193), 19 (MTO207), 5 (MTO219), 14 (MTO34), 16 (MTO68) and 29 (MTO93). Right, distribution of tumour diameters.
Extended Data Figure 7 Source of tumoral TGFβ and pathway blockade in vivo.
a, b, TGFB1−TGFB3 (a) or Tgfb1–Tgfb3 (b) mRNA expression levels in sorted cell populations from disaggregated human CRCs (a) or mouse liver metastasis (b). First, epithelial cancer cells, immune cells, endothelial cells and CAFs were sorted with labelled antibodies against EPCAM, CD45 (n = 14 patients for each), CD31 and FAP (n = 6 for each) for human and PDGFRB for mouse. Subsequently, RNA from sorted cells was extracted and analysed by microarray (human) or reverse transcription–quantitative PCR (RT–qPCR) (mouse). a, Standardized expression data are shown as z score. Individual values and Tukey box plots; P values are for TGFB1: epithelial versus leukocytes, 7.5 × 10−6; epithelial versus endothelial, 6.2 × 10−4; epithelial versus CAFs, 6.2 × 10−4; and the Kruskal–Wallis (KW) P value is 6.3 × 10−6. For TGFB2: CAFs versus epithelial, 6.2 × 10−4; CAFs versus leukocytes, 8.4 × 10−4; CAFs versus endothelial, 0.0051; the Kruskal–Wallis P value is 0.0017. For TGFB3: epithelial versus CAFs, 6.2 × 10−4; epithelial versus leukocyte, 0.0409; epithelial versus endothelial, 0.0150; leukocyte versus CAFs, 6.2 × 10−4; endothelial versus CAFs, 0.0051; the Kruskal–Wallis P value is 8.5 × 10−5. Direct comparisons using two-sided Mann–Whitney U test. Data are shown as mRNA expression levels normalized to epithelial cells, mean ± s.e.m. from technical triplicates; Tgfb2 was undetectable (ND) in leukocytes. Note that in both settings, TGFB1 is expressed by all stromal cell types, TGFB2 and TGFB3 are mainly expressed by CAFs. Expression in epithelial cancer cells is comparatively low. c–h, Immunohistochemistry for TGFβ target gene products in the TME of MTO138-derived liver metastases, representing two independent experiments (c, e, g), as well as for the inhibition of stromal TGFβ signalling by galunisertib (d, f, h). c, d, Micrographs of sections of liver metastases stained for pSMAD2. e, f, Staining reveals expression of the TGFβ target CALD1 exclusively in the metastatic TME. Treatment with galunisertib for three days abrogates expression of this cytoplasmic protein. g, h, Staining for the TGFβ target IGFBP7, which is exclusively expressed in the metastatic TME. Treatment with galunisertib for three days abrogates expression of this secreted protein. Scale bars, 100 μm.
Extended Data Figure 8 Characterization of MTOs in metastatic experiments.
a, Representative examples of GFP positivity measured by flow cytometry, indicating genetic recombination in the UBCcreERT2;Tgfbr2fl/fl genetic model (representative of three independent experiments). b, Liver metastases counted five weeks after intrasplenic injection of LAKP MTO54 or MTO220 and treatment from day 2 to day 14 with galunisertib (red; n = 4 mice for MTO54 and n = 10 for MTO220) or control (blue; n = 5 mice for MTO54 and n = 10 for MTO220), mean ± s.e.m., 95% confidence interval for MTO220 is (−3–0), from a two-sided Mann–Whitney U test. c, Stromal TGFβ activation markers in LAKP MTO liver tumours, both supporting the classification of these tumours as CMS4 (Fig. 1i) and explaining the efficacy of galunisertib in b; representative of two independent experiments. d, Number of nodules five weeks after injection of LAKP empty guide or LAKPS (LAKP + S) MTOs and treatment with galunisertib (red) or control (blue), mean ± s.e.m., 95% confidence interval for LAKPS is (−554 to −176), n = 5 mice per condition; two-sided Mann–Whitney U test. e, Number of liver nodules (mean ± s.e.m.) four weeks after intrasplenic injection of MTOs (either unlabelled or with mCherry–luciferase vector, or after CRISPR knockout of the mCherry–luciferase coding region for MTO138), after treatment. 95% confidence intervals are: for MTO93, (−113−0), n = 5 mice per condition; MTO93-LUC: (−104 to −4), n = 5 control and n = 4 galunisertib; MTO140, (−297 to −21), n = 7 control and n = 5 galunisertib; MTO140-LUC, (−251 to −21), n = 5 per condition; MTO138-LUC, (−86 to −8), n = 5 per condition; MTO138-KO-LUC, (−70 to −13), n = 7 per condition; two-sided Mann–Whitney U test. f, Longitudinal intravital bioluminescence imaging (BLI) quantification (photons s−1, normalized to day 0) of an intrasplenic metastasis initiation experiment with MTO140 in C57BL/6J mice treated with galunisertib or control until day 14, n = 5 mice per condition. Points and lines represent individual mice, trend lines (bold) show a LOESS model with 95% confidence interval (grey band). 95% confidence interval for the difference at day 25: (−222.7 to −23.9), two-sided Mann–Whitney U test. g, BLI (as in f) of a metastasis-initiation experiment with MTO138 in C57BL/6J mice, n = 24 control mice, n = 15 galunisertib. h, BLI (as in f) of galunisertib treatment in intrasplenic liver colonization assays with MTO129 or MTO138, in C57BL/6J (n = 7 mice per condition, except for MTO138 + Gal, n = 11) or athymic nu/nu mice (n = 5 per condition). Note the complete absence of treatment effect in the mice lacking functional T cells. i, Number of liver nodules (mean ± s.e.m.) in mice intrasplenically injected with MTO129 and treated with galunisertib alone or with antibodies against CD4 or IgG control. 95% confidence interval for galunisertib versus control is (−79 to −8), n = 5 mice per condition; two-sided Mann–Whitney U test. j, Number of liver nodules (mean ± s.e.m.) after intrasplenic injection of MTO93 in syngeneic, Lgr5eGFP-creERT2 mice—from the colonies that gave rise to the 3× and 4× genetic models—treated with galunisertib (red) or control (blue). 95% confidence interval is (−13 to −1), n = 20 control mice and n = 25 for galunisertib; two-sided Mann–Whitney U test. Scale bars, 500 μm.
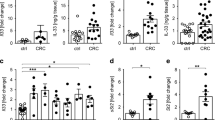
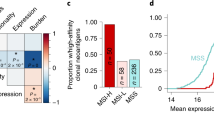
Extended Data Figure 9 Immune modulation by TGFβ in CRC.
A human meta-cohort was used to analyse the interaction of TGFβ expression levels and T-cell-activation state. a, Expression signature ratio between active CD4 TH1 and naive CD4 T cells differs significantly between MSS and MSI patients. Standardized expression for individual patients and Tukey box plots; MSS, n = 981 samples; MSI, n = 198; two-sided Wald test. b, c, Expression signature for naive CD4 T cells (b) and TH1-differentiated CD4 T cells (c) in patients with low, medium and high expression of TGFB1–TGFB3. Standardized expression for individual patients and Tukey box plots. P values (b): low versus medium (n = 327), 1.7 × 10−4; low versus high, 2.2 × 10−16; medium versus high, 2.2 × 10−16. P values (c): low versus medium, 0.0503; low versus high, 6.8 × 10−14; medium versus high, 8.1 × 10−8; n = 327 patient samples per group; two-sided Wald test. The log-likelihood ratio test P value is 2.2 × 10−16 for b and 2.3 × 10−14 for c. d, TGFB1–3 mRNA expression levels predict poor prognosis. Hazard risk (HR) 95% confidence intervals are: low versus medium, (0.65–1.42); low versus high, (1.08–2.20); medium versus high, (1.12–2.32); n = 255 patient samples (low), n = 254 (medium) and n = 249 (high); two-sided Wald test. e, f, Correlation between ratio of CD4 TH1 versus naive CD4 signatures and TGFB1–3 mRNA expression levels (e) or expression of a FAP+ CAF signature (f) in n = 981 MSS patient samples. Pearson correlation coefficients are indicated; two-sided Wald test. g, Kaplan–Meier curves for relapse-free survival for patients according CD4 TH1-naive signatures ratio. Hazard risk 95% confidence intervals are: low versus medium (0.40–0.81), low versus high (0.37–0.78), medium versus high (0.64–1.41); n = 243 (low), n = 259 (medium) and n = 257 (high); two-sided Wald test. h, Cell densities in micrometastases 10 days after injection, treated with vehicle (control, blue) or galunisertib (red); shown are individual MTO129 and MTO140 tumour values and mouse means (diamonds and triangles, respectively), together with group mean ± s.e.m. derived from a mixed-effects linear model; n = 4 mice per condition (MTO129) and n = 6 for MTO140. i, Representative images of immunohistochemistry of the MTO140 micrometastases quantified in h. Mean cell densities (cells mm−2) are indicated. Images represent two independent experiments. Scale bars, 100 μm.
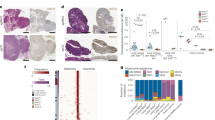
Extended Data Figure 10 TGFβ inhibition and the tumour immune microenvironment.
a–d, Densities of tumour-infiltrating lymphocytes in galunisertib (Gal, red) or control (blue) treated liver metastases derived from primary LAKTP MTOs (a), in treated orthotopically injected MTO-derived primary cancers (b), in liver metastases in the UBCcreERT2;Tgfbr2fl/fl background compared to UBC+/+;Tgfbr2fl/fl control (c), or two days after start of treatment in established liver metastases (d; treatment started on day 14). Shown are individual tumours (grey circles, a, c, d), mouse means (squares, or triangles for MTO93, a), and group means ± s.e.m. Data are analysed with a mixed-effects linear model; in (a) n = 5 (control) or n = 4 (galunisertib) mice per condition; in (b), n = 9 (control) or n = 8 (galunisertib) mice per condition; in (c), n = 5 (UBC+/+) or n = 7 (UBC creERT2 ;Tgfbr2 fl/fl) mice per condition; and in (d), n = 6 mice per condition. e, Distribution of cell types within the PD-L1+ population (mean ± s.e.m.) in microdissected tumours (euthanized at day 18, four days after treatment start); n = 3 mice. f, Myeloid cell types (gated for CD45/(CD11b and/or CD11c)) within the CD45+ PD-L1+ population. Mean ± s.e.m. percentage of n = 3 mice.
Supplementary information
Supplementary Information
This file contains the uncropped western blots, a Supplementary Discussion, Supplementary Acknowledgements, Supplementary Methods and Supplementary References (PDF 3487 kb)
Source data
Rights and permissions
About this article
Cite this article
Tauriello, D., Palomo-Ponce, S., Stork, D. et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 554, 538–543 (2018). https://doi.org/10.1038/nature25492
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25492
This article is cited by
-
Macrophage heterogeneity and its interactions with stromal cells in tumour microenvironment
Cell & Bioscience (2024)
-
Transforming growth factor-induced gene TGFBI is correlated with the prognosis and immune infiltrations of breast cancer
World Journal of Surgical Oncology (2024)
-
INHBA(+) cancer-associated fibroblasts generate an immunosuppressive tumor microenvironment in ovarian cancer
npj Precision Oncology (2024)
-
Multi-omic analyses of m5C readers reveal their characteristics and immunotherapeutic proficiency
Scientific Reports (2024)
-
SOX17 enables immune evasion of early colorectal adenomas and cancers
Nature (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.