Abstract
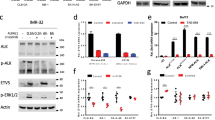
Neuroblastoma, an embryonal tumour of the peripheral sympathetic nervous system, accounts for approximately 15% of all deaths due to childhood cancer1. High-risk neuroblastomas are rapidly progressive; even with intensive myeloablative chemotherapy, relapse is common and almost uniformly fatal2,3. Here we report the detection of previously unknown mutations in the ALK gene, which encodes a receptor tyrosine kinase, in 8% of primary neuroblastomas. Five non-synonymous sequence variations were identified in the kinase domain of ALK, of which three were somatic and two were germ line. The most frequent mutation, F1174L, was also identified in three different neuroblastoma cell lines. ALK complementary DNAs encoding the F1174L and R1275Q variants, but not the wild-type ALK cDNA, transformed interleukin-3-dependent murine haematopoietic Ba/F3 cells to cytokine-independent growth. Ba/F3 cells expressing these mutations were sensitive to the small-molecule inhibitor of ALK, TAE684 (ref. 4). Furthermore, two human neuroblastoma cell lines harbouring the F1174L mutation were also sensitive to the inhibitor. Cytotoxicity was associated with increased amounts of apoptosis as measured by TdT-mediated dUTP nick end labelling (TUNEL). Short hairpin RNA (shRNA)-mediated knockdown of ALK expression in neuroblastoma cell lines with the F1174L mutation also resulted in apoptosis and impaired cell proliferation. Thus, activating alleles of the ALK receptor tyrosine kinase are present in primary neuroblastoma tumours and in established neuroblastoma cell lines, and confer sensitivity to ALK inhibition with small molecules, providing a molecular rationale for targeted therapy of this disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
Microarray data have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) public database. The accession numbers for the SNP array analyses are GSM206563 and GSM206564.
References
National Cancer Institute Surveillance, Epidemiology and End Results Database. <http://seer.cancer.gov> (2005)
Matthay, K. K. et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children’s Cancer Group. N. Engl. J. Med. 341, 1165–1173 (1999)
George, R. E. et al. High risk neuroblastoma treated with tandem autologous peripheral-blood stem cell-supported transplant: long-term survival update. J. Clin. Oncol. 24, 2891–2896 (2006)
Galkin, A. V. et al. Identification of NVP-TAE684, a potent, selective, and efficacious inhibitor of NPM-ALK. Proc. Natl Acad. Sci. USA 104, 270–275 (2007)
George, R. E. et al. Genome-wide analysis of neuroblastomas using high-density single nucleotide polymorphism arrays. PLoS One 2, e255–264 (2007)
Morris, S. W. et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science 263, 1281–1284 (1994)
Soda, M. et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 (2007)
Rikova, K. et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 (2007)
Chiarle, R., Voena, C., Ambrogio, C., Piva, R. & Inghirami, G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nature Rev. Cancer 8, 11–23 (2008)
The International HapMap Consortium. The International HapMap Project. Nature 426, 789–796 (2003)
Greulich, H. et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2, e313 (2005)
Stephens, P. et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature 431, 525–526 (2004)
Lynch, T. J. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139 (2004)
Paez, J. G. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004)
Jiang, J. et al. Epidermal growth factor-independent transformation of Ba/F3 cells with cancer-derived epidermal growth factor receptor mutants induces gefitinib-sensitive cell cycle progression. Cancer Res. 65, 8968–8974 (2005)
Fröhling, S. et al. Identification of driver and passenger mutations of FLT3 by high-throughput DNA sequence analysis and functional assessment of candidate alleles. Cancer Cell 12, 501–513 (2007)
Koivunen, J. P. et al. EML4–ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin. Cancer Res. 14, 4275–4283 (2008)
Li, R. & Morris, S. W. Development of anaplastic lymphoma kinase (ALK) small-molecule inhibitors for cancer therapy. Med. Res. Rev. 28, 372–412 (2008)
McDermott, U. et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 68, 3389–3395 (2008)
Lamant, L. et al. Expression of the ALK tyrosine kinase gene in neuroblastoma. Am. J. Pathol. 156, 1711–1721 (2000)
Miyake, I. et al. Activation of anaplastic lymphoma kinase is responsible for hyperphosphorylation of ShcC in neuroblastoma cell lines. Oncogene 21, 5823–5834 (2002)
Osajima-Hakomori, Y. et al. Biological role of anaplastic lymphoma kinase in neuroblastoma. Am. J. Pathol. 167, 213–222 (2005)
Stommel, J. M. et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 318, 287–290 (2007)
Engelman, J. A. et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043 (2007)
Nakagawara, A. et al. Differential expression of pleiotrophin and midkine in advanced neuroblastomas. Cancer Res. 55, 1792–1797 (1995)
Calvet, L. et al. Pleiotrophin, a candidate gene for poor tumor vasculature and in vivo neuroblastoma sensitivity to irinotecan. Oncogene 25, 3150–3159 (2006)
Nickerson, D. A., Tobe, V. O. & Taylor, S. L. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 25, 2745–2751 (1997)
Acknowledgements
We thank J. Elechko and L. Moreau for technical assistance, A. Azarova for help with cell culture and cell growth assays, Q. Jiang and X. Cui for assistance during ALK small-molecule inhibitor development, and A. Kung for Ba/F3 cells and retroviral vectors. We acknowledge the Children’s Oncology Group for provision of neuroblastoma tumour and matched normal DNAs, and tumour touch prep slides. We thank J. Maris for neuroblastoma cell lines. We thank Abbott Molecular International for the Vysis LSI ALK Dual Color, Break Apart Rearrangement Probe. This work was supported by grants from the National Institutes of Health (R.E.G.), the Friends for Life Neuroblastoma Fund (R.E.G.), the Children’s Oncology Group (R.E.G.), Alex’s Lemonade Stand Foundation (M.H. and M.M.), NCI CA69129 (L.X. and S.W.M.), Cancer Center Core grant CA21765 (T.R.W., L.X. and S.W.M.), the American Lebanese Syrian Associated Charities and St. Jude Children’s Research Hospital (T.R.W., L.X. and S.W.M.).
Author Contributions R.E.G., M.M. and A.T.L. designed the experiments and wrote the manuscript. M.H., H.G. and M.M. performed the DNA sequencing and analysis. T.S., S.F., W.L., Y.A., H.G. and R.E.G. carried out the functional analyses. J.Z., W.Z. and N.S.G. performed the homology modelling and synthesis of TAE684. W.B.L. and P.M. performed the statistical analysis. S.Z., V.E.G. and T.R.W. were involved with the design of ALK inhibitors. L.X. and S.W.M. assisted with experimental design, provided reagents and advice on ALK inhibitors. M.M., L.D. and D.G.G. provided advice on the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-3 with Legends, Supplementary Methods and a Supplementary Table 1. (PDF 790 kb)
Rights and permissions
About this article
Cite this article
George, R., Sanda, T., Hanna, M. et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature 455, 975–978 (2008). https://doi.org/10.1038/nature07397
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature07397
This article is cited by
-
Comprehensive exploration of the involvement of cuproptosis in tumorigenesis and progression of neuroblastoma
BMC Genomics (2023)
-
Genomic ALK alterations in primary and relapsed neuroblastoma
British Journal of Cancer (2023)
-
Circulating tumor DNA reveals mechanisms of lorlatinib resistance in patients with relapsed/refractory ALK-driven neuroblastoma
Nature Communications (2023)
-
Lorlatinib with or without chemotherapy in ALK-driven refractory/relapsed neuroblastoma: phase 1 trial results
Nature Medicine (2023)
-
Recent advances in the developmental origin of neuroblastoma: an overview
Journal of Experimental & Clinical Cancer Research (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.