Abstract
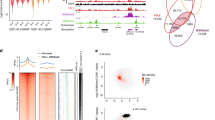
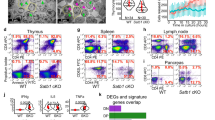
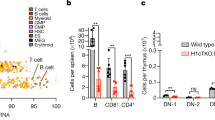
Eukaryotic chromosomes are organized inside the nucleus in such a way that only a subset of the genome is expressed in any given cell type, but the details of this organization are largely unknown1,2,3. SATB1 (‘special AT-rich sequence binding 1’), a protein found predominantly in thymocytes4, regulates genes by folding chromatin into loop domains, tethering specialized DNA elements to an SATB1 network structure5. Ablation of SATB1 by gene targeting results in temporal and spatial mis-expression of numerous genes and arrested T-cell development, suggesting that SATB1 is a cell-type specific global gene regulator6. Here we show that SATB1 targets chromatin remodelling to the IL-2Rα (‘interleukin-2 receptor α’) gene, which is ectopically transcribed in SATB1 null thymocytes. SATB1 recruits the histone deacetylase contained in the NURD chromatin remodelling complex to a SATB1-bound site in the IL-2Rα locus, and mediates the specific deacetylation of histones in a large domain within the locus. SATB1 also targets ACF1 and ISWI, subunits of CHRAC and ACF nucleosome mobilizing complexes, to this specific site and regulates nucleosome positioning over seven kilobases. SATB1 defines a class of transcriptional regulators that function as a ‘landing platform’ for several chromatin remodelling enzymes and hence regulate large chromatin domains.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Felsenfeld, G. Chromatin unfolds. Cell 86, 13–19 (1996)
Travers, A. An engine for nucleosome remodeling. Cell 96, 311–314 (1999)
Bulger, M. & Groudine, M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 13, 2465–2477 (1999)
Dickinson, L. A., Joh, T., Kohwi, Y. & Kohwi-Shigematsu, T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell 70, 631–645 (1992)
Cai, S., Han, H.-J. & Kohwi-Shigematsu, T. SATB1 creates chromatin domains with specific histone modifications to regulate genes. Cell (submitted)
Alvarez, J. D. et al. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 14, 521–535 (2000)
Kohwi-Shigematsu, T. & Kohwi, Y. Torsional stress stabilizes extended base unpairing in suppressor sites flanking immunoglobulin heavy chain enhancer. Biochemistry 29, 9551–9560 (1990)
Bode, J. et al. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science 255, 195–197 (1992)
Kingston, R. E. & Narlikar, G. J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13, 2339–2352 (1999)
Varga-Weisz, P. ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene 20, 3076–3085 (2001)
Fry, C. J. & Peterson, C. L. Chromatin remodeling enzymes: who's on first? Curr. Biol. 11, R185–R197 (2001)
Dickinson, L. A. & Kohwi-Shigematsu, T. Nucleolin is a matrix attachment region DNA-binding protein that specifically recognizes a region with high base-unpairing potential. Mol. Cell Biol. 15, 456–465 (1995)
Knoepfler, P. S. & Eisenman, R. N. Sin meets NuRD and other tails of repression. Cell 99, 447–450 (1999)
Zhang, Y., LeRoy, G., Seelig, H. P., Lane, W. S. & Reinberg, D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95, 279–289 (1998)
Taunton, J., Hassig, C. A. & Schreiber, S. L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–411 (1996)
Xue, Y. et al. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2, 851–861 (1998)
Langst, G. & Becker, P. B. Nucleosome mobilization and positioning by ISWI-containing chromatin- remodeling factors. J. Cell Sci. 114, 2561–2568 (2001)
Galande, S. & Kohwi-Shigematsu, T. Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J. Biol. Chem. 274, 20521–20528 (1999)
Poot, R. A. et al. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 19, 3377–3387 (2000)
de Belle, I., Cai, S. & Kohwi-Shigematsu, T. The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J. Cell Biol. 141, 335–348 (1998)
Kohwi-Shigematsu, T., deBelle, I., Dickinson, L. A., Galande, S. & Kohwi, Y. Identification of base-unpairing region-binding proteins and characterization of their in vivo binding sequences. Methods Cell Biol. 53, 323–354 (1998)
Kouzarides, T. Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev. 9, 40–48 (1999)
Roth, S. Y., Denu, J. M. & Allis, C. D. Histone acetyltransferases. Annu. Rev. Biochem. 70, 81–120 (2001)
Soldaini, E. et al. Mouse interleukin-2 receptor alpha gene expression. Delimitation of cis-acting regulatory elements in transgenic mice and by mapping of DNase-I hypersensitive sites. J. Biol. Chem. 270, 10733–10742 (1995)
Fernandez, L. A., Winkler, M. & Grosschedl, R. Matrix attachment region-dependent function of the immunoglobulin µ enhancer involves histone acetylation at a distance without changes in enhancer occupancy. Mol. Cell Biol. 21, 196–208 (2001)
Kirillov, A. et al. A role for nuclear NF-κB in B-cell-specific demethylation of the Igκ locus. Nature Genet. 13, 435–441 (1996)
Lichtenstein, M., Keini, G., Cedar, H. & Bergman, Y. B cell-specific demethylation: a novel role for the intronic κ chain enhancer sequence. Cell 76, 913–923 (1994)
Jenuwein, T. et al. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature 385, 269–272 (1997)
Forrester, W. C., Fernandez, L. A. & Grosschedl, R. Nuclear matrix attachment regions antagonize methylation-dependent repression of long-range enhancer-promoter interactions. Genes Dev. 13, 3003–3014 (1999)
Collins, N. et al. An ACF1–ISWI nucleosome remodelling complex is required for DNA replication through condensed chromatin in mammalian cells. Nature Genet. (in the press)
Acknowledgements
We thank the Joint Genome Institute, especially K. Kadner, for sequencing the BAC clone containing the IL-2Rα locus; Y. Zhang for antibodies against Mi-2 and MTA-2; H.-J. Han and S. Galande for DNA constructs; M. Grimaldi and I. Kukimoto for expressing recombinant SNF2H and hACF1; and C. García-Jiménez for affinity purification of antibodies. We also thank C. Goding and M. Kohwi for comments on the manuscript. This work was supported by the National Institutes of Health (T.K-S.), the Marie Curie Cancer Care foundation (P.V-W.) and training grant AG00266 from the National Institute on Aging (D.Y.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Yasui, D., Miyano, M., Cai, S. et al. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419, 641–645 (2002). https://doi.org/10.1038/nature01084
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature01084
This article is cited by
-
A dual function for the chromatin organizer Special A-T rich Binding Protein 1 in B-lineage cells
Cellular & Molecular Immunology (2023)
-
HDAC5 modulates SATB1 transcriptional activity to promote lung adenocarcinoma
British Journal of Cancer (2023)
-
The role of transcription factors in shaping regulatory T cell identity
Nature Reviews Immunology (2023)
-
Expression of SATB1 and SATB2 in the brain of bony fishes: what fish reveal about evolution
Brain Structure and Function (2023)
-
Long Non-Coding RNA ANRIL Regulates Inflammatory Factor Expression in Ulcerative Colitis Via the miR-191-5p/SATB1 Axis
Inflammation (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.