Abstract
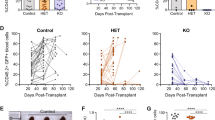
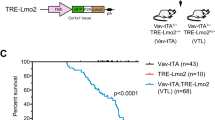
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematopoietic neoplasm resulting from the malignant transformation of T-cell progenitors, and comprises ~15% and 25% of pediatric and adult ALL cases, respectively. It is well-established that activating NOTCH1 mutations are the major genetic lesions driving T-ALL in most patients, but efforts to develop targeted therapies against this pathway have produced limited success in decreasing leukemic burden and come with significant clinical side effects. A finer detailed understanding of the genetic and molecular mechanisms underlying T-ALL is required identify patients at increased risk for treatment failure and the development of precision medicine strategies. Generation of genetic models that more accurately reflect the normal developmental history of T-ALL are necessary to identify new avenues for treatment. The DNA methyltransferase enzyme DNMT3A is also recurrently mutated in T-ALL patients, and we show here that inactivation of Dnmt3a combined with Notch1 gain-of-function leads to an aggressive T-ALL in mouse models. Moreover, conditional inactivation of Dnmt3a in mouse hematopoietic cells leads to an accumulation of immature progenitors in the thymus, which are less apoptotic. These data demonstrate that Dnmt3a is required for normal T-cell development, and acts as a T-ALL tumor suppressor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Aifantis I, Raetz E, Buonamici S . Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol 2008; 8: 380–390.
Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 1999; 10: 547–558.
Kopan R, Ilagan MX . The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 2009; 137: 216–233.
Huang YH, Li D, Winoto A, Robey EA . Distinct transcriptional programs in thymocytes responding to T cell receptor, Notch, and positive selection signals. Proc Natl Acad Sci USA 2004; 101: 4936–4941.
Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A . Signalling downstream of activated mammalian Notch. Nature 1995; 377: 355–358.
Reizis B, Leder P . Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev 2002; 16: 295–300.
Weng AP, Ferrando AA, Lee W, JPt Morris, Silverman LB, Sanchez-Irizarry C et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271.
Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med 1996; 183: 2283–2291.
Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE et al. DNMT3A mutations in acute myeloid leukemia". N Engl J Med 2010; 363: 2424–2433.
Cancer Genome Atlas Research. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–2074.
Simon C, Chagraoui J, Krosl J, Gendron P, Wilhelm B, Lemieux S et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev 2012; 26: 651–656.
Aref S, El Menshawy N, El-Ghonemy MS, Zeid TA, El-Baiomy MA . Clinicopathologic effect of DNMT3A mutation in adult T-cell acute lymphoblastic leukemia. Clin Lymphoma Myeloma Leuk 2016; 16: 43–48.
Couronne L, Bastard C, Bernard OA . TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med 2012; 366: 95–96.
Grossmann V, Haferlach C, Weissmann S, Roller A, Schindela S, Poetzinger F et al. The molecular profile of adult T-cell acute lymphoblastic leukemia: mutations in RUNX1 and DNMT3A are associated with poor prognosis in T-ALL. Genes Chromosomes Cancer 2013; 52: 410–422.
Van Vlierberghe P, Ambesi-Impiombato A, Perez-Garcia A, Haydu JE, Rigo I, Hadler M et al. ETV6 mutations in early immature human T cell leukemias. J Exp Med 2011; 208: 2571–2579.
Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS et al. "Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 2012; 44: 23–31.
Challen GA, Goodell MA . Runx1 isoforms show differential expression patterns during hematopoietic development but have similar functional effects in adult hematopoietic stem cells. Exp Hematol 2010; 38: 403–416.
Chiang MY, Xu L, Shestova O, Histen G, L'Heureux S, Romany C et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest 2008; 118: 3181–3194.
Challen GA, Sun D, Mayle A, Jeong M, Luo M, Rodriguez B et al. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell 2014; 15: 350–364.
Akalin A, Garrett-Bakelman FE, Kormaksson M, Busuttil J, Zhang L, Khrebtukova I et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet 2012; 8: e1002781.
Mayle A, Yang L, Rodriguez B, Zhou T, Chang E, Curry CV et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood 2015; 125: 629–638.
Yang L, Rodriguez B, Mayle A, Park HJ, Lin X, Luo M et al. DNMT3A loss drives enhancer hypomethylation in FLT3-ITD-associated leukemias. Cancer Cell 2016; 29: 922–934.
Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012; 481: 157–163.
Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol 2009; 10: 147–156.
Neumann M, Heesch S, Schlee C, Schwartz S, Gokbuget N, Hoelzer D et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood 2013; 121: 4749–4752.
Murtaugh LC, Stanger BZ, Kwan KM, Melton DA . Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci USA 2003; 100: 14920–14925.
Calnan BJ, Szychowski S, Chan FK, Cado D, Winoto A . A role for the orphan steroid receptor Nur77 in apoptosis accompanying antigen-induced negative selection. Immunity 1995; 3: 273–282.
Zhou T, Cheng J, Yang P, Wang Z, Liu C, Su X et al. Inhibition of Nur77/Nurr1 leads to inefficient clonal deletion of self-reactive T cells. J Exp Med 1996; 183: 1879–1892.
Lin B, Kolluri SK, Lin F, Liu W, Han YH, Cao X et al. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell 2004; 116: 527–540.
Thompson J, Winoto A . During negative selection, Nur77 family proteins translocate to mitochondria where they associate with Bcl-2 and expose its proapoptotic BH3 domain. J Exp Med 2008; 205: 1029–1036.
Wang A, Rud J, Olson CM Jr., Anguita J, Osborne BA . Phosphorylation of Nur77 by the MEK-ERK-RSK cascade induces mitochondrial translocation and apoptosis in T cells. J Immunol 2009; 183: 3268–3277.
Lee SL, Wesselschmidt RL, Linette GP, Kanagawa O, Russell JH, Milbrandt J . Unimpaired thymic and peripheral T cell death in mice lacking the nuclear receptor NGFI-B (Nur77). Science 1995; 269: 532–535.
Schmitt TM, Zuniga-Pflucker JC . Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 2002; 17: 749–756.
Zhan Y, Du X, Chen H, Liu J, Zhao B, Huang D et al. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat Chem Biol 2008; 4: 548–556.
Celik H, Mallaney C, Kothari A, Ostrander EL, Eultgen E, Martens A et al. Enforced differentiation of Dnmt3a-null bone marrow leads to failure with c-Kit mutations driving leukemic transformation. Blood 2015; 125: 619–628.
Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 2014; 25: 442–454.
Kim SJ, Zhao H, Hardikar S, Singh AK, Goodell MA, Chen T . A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood 2013; 122: 4086–4089.
Spencer DH, Russler-Germain DA, Ketkar S, Helton NM, Lamprecht TL, Fulton RS et al. CpG island hypermethylation mediated by DNMT3A is a consequence of AML progression. Cell 2017; 168: 801–816.
Scourzic L, Couronne L, Pedersen MT, Della Valle V, Diop M, Mylonas E et al. DNMT3A(R882H) mutant and Tet2 inactivation cooperate in the deregulation of DNA methylation control to induce lymphoid malignancies in mice. Leukemia 2016; 30: 1388–1398.
Koya J, Kataoka K, Sato T, Bando M, Kato Y, Tsuruta-Kishino T et al. DNMT3A R882 mutants interact with polycomb proteins to block haematopoietic stem and leukaemic cell differentiation. Nature Commun 2016; 7: 10924.
Thomas DA, Kantarjian H, Smith TL, Koller C, Cortes J, O'Brien S et al. Primary refractory and relapsed adult acute lymphoblastic leukemia: characteristics, treatment results, and prognosis with salvage therapy. Cancer 1999; 86: 1216–1230.
Acknowledgements
We thank the Alvin J Siteman Cancer Center at Washington University School of Medicine for the use of the Siteman Flow Cytometry Core, which provided cell sorting and analysis. The Siteman Cancer Center is supported in part by NCI Cancer Center Support Grant CA91842. We thank the Genome Technology Access Center Washington University School of Medicine for genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant CA91842 and by ICTS/CTSA Grant UL1TR000448 NIH, and NIH Roadmap for Medical Research. Research reported in this publication was supported by the Washington University Institute of Clinical and Translational Sciences grant UL1TR000448 NIH. The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIH. This work was supported by grants (to GAC) from Alex’s Lemonade Stand Foundation, the Children’s Discovery Institute (MC-II-2013-286), the American Society of Hematology, and the Sidney Kimmel Foundation. WCW was supported by NIH T32HL008088. HC was supported by an American Society of Hematology scholar award. JN was supported in part by an Alex’s Lemonade Stand Foundation POST award. CM was supported in part by NIH T32HL008088, and NIH DK111058-01. ELO was supported by NIH 5T32CA113275-10.
Author contributions
Designed and performed experiments: ACK, AK, WCW, HC, JN, CM, EE, AM, GAC. Analyzed data: ELO, MCV, ALY, TED, MEF, BZ, GAC. Supervised the study and wrote the paper: GAC.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Kramer, A., Kothari, A., Wilson, W. et al. Dnmt3a regulates T-cell development and suppresses T-ALL transformation. Leukemia 31, 2479–2490 (2017). https://doi.org/10.1038/leu.2017.89
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.89
This article is cited by
-
T Cell Repertoire Abnormality in Immunodeficiency Patients with DNA Repair and Methylation Defects
Journal of Clinical Immunology (2022)
-
Clonal hematopoiesis and its emerging effects on cellular therapies
Leukemia (2021)
-
Functional and epigenetic phenotypes of humans and mice with DNMT3A Overgrowth Syndrome
Nature Communications (2021)
-
DNA methylation disruption reshapes the hematopoietic differentiation landscape
Nature Genetics (2020)
-
Lymphopenia at diagnosis is highly prevalent in myelodysplastic syndromes and has an independent negative prognostic value in IPSS-R-low-risk patients
Blood Cancer Journal (2019)