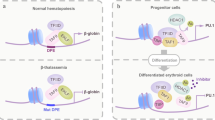
Abstract
Hematopoiesis is coordinated by a complex regulatory network of transcription factors and among them PU.1 (Spi1, Sfpi1) represents a key molecule. This review summarizes the indispensable requirement of PU.1 during hematopoietic cell fate decisions and how the function of PU.1 can be modulated by protein–protein interactions with additional factors. The mutual negative regulation between PU.1 and GATA-1 is detailed within the context of normal and leukemogenic hematopoiesis and the concept of ‘differentiation therapy’ to restore normal cellular differentiation of leukemic cells is discussed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C et al. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev 1997; 11: 774–785.
Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 2006; 126: 755–766.
Cross MA, Enver T . The lineage commitment of haemopoietic progenitor cells. Curr Opin Genet Dev 1997; 7: 609–613.
Krysinska H, Hoogenkamp M, Ingram R, Wilson N, Tagoh H, Laslo P et al. A two-step, PU.1-dependent mechanism for developmentally regulated chromatin remodeling and transcription of the c-fms gene. Mol Cell Biol 2007; 27: 878–887.
Kodandapani R, Pio F, Ni CZ, Piccialli G, Klemsz M, McKercher S et al. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature 1996; 380: 456–460.
Burda P, Curik N, Kokavec J, Basova P, Mikulenkova D, Skoultchi AI et al. PU.1 activation relieves GATA-1-mediated repression of Cebpa and Cbfb during leukemia differentiation. Mol Cancer Res 2009; 7: 1693–1703.
Gangenahalli GU, Gupta P, Saluja D, Verma YK, Kishore V, Chandra R et al. Stem cell fate specification: role of master regulatory switch transcription factor PU.1 in differential hematopoiesis. Stem Cells Dev 2005; 14: 140–152.
Weigelt K, Lichtinger M, Rehli M, Langmann T . Transcriptomic profiling identifies a PU.1 regulatory network in macrophages. Biochem Biophys Res Commun 2009; 380: 308–312.
Ross IL, Yue X, Ostrowski MC, Hume DA . Interaction between PU.1 and another Ets family transcription factor promotes macrophage-specific Basal transcription initiation. J Biol Chem 1998; 273: 6662–6669.
Eichbaum QG, Iyer R, Raveh DP, Mathieu C, Ezekowitz RA . Restriction of interferon gamma responsiveness and basal expression of the myeloid human Fc gamma R1b gene is mediated by a functional PU.1 site and a transcription initiator consensus. J Exp Med 1994; 179: 1985–1996.
Hoogenkamp M, Krysinska H, Ingram R, Huang G, Barlow R, Clarke D et al. The Pu.1 locus is differentially regulated at the level of chromatin structure and noncoding transcription by alternate mechanisms at distinct developmental stages of hematopoiesis. Mol Cell Biol 2007; 27: 7425–7438.
Petrovick MS, Hiebert SW, Friedman AD, Hetherington CJ, Tenen DG, Zhang DE . Multiple functional domains of AML1: PU.1 and C/EBPalpha synergize with different regions of AML1. Mol Cell Biol 1998; 18: 3915–3925.
Hagemeier C, Bannister AJ, Cook A, Kouzarides T . The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc Natl Acad Sci USA 1993; 90: 1580–1584.
Rehli M, Niller HH, Ammon C, Langmann S, Schwarzfischer L, Andreesen R et al. Transcriptional regulation of CHI3L1, a marker gene for late stages of macrophage differentiation. J Biol Chem 2003; 278: 44058–44067.
Liu J, Ma X . Interferon regulatory factor 8 regulates RANTES gene transcription in cooperation with interferon regulatory factor-1, NF-kappaB, and PU.1. J Biol Chem 2006; 281: 19188–19195.
Heydemann A, Juang G, Hennessy K, Parmacek MS, Simon MC . The myeloid-cell-specific c-fes promoter is regulated by Sp1, PU.1, and a novel transcription factor. Mol Cell Biol 1996; 16: 1676–1686.
Eisenbeis CF, Singh H, Storb U . PU.1 is a component of a multiprotein complex which binds an essential site in the murine immunoglobulin lambda 2-4 enhancer. Mol Cell Biol 1993; 13: 6452–6461.
Pongubala JM, Nagulapalli S, Klemsz MJ, McKercher SR, Maki RA, Atchison ML . PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3′ enhancer activity. Mol Cell Biol 1992; 12: 368–378.
Back J, Allman D, Chan S, Kastner P . Visualizing PU.1 activity during hematopoiesis. Exp Hematol 2005; 33: 395–402.
Nutt SL, Metcalf D, D’Amico A, Polli M, Wu L . Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med 2005; 201: 221–231.
Chen H, Ray-Gallet D, Zhang P, Hetherington CJ, Gonzalez DA, Zhang DE et al. PU.1 (Spi-1) autoregulates its expression in myeloid cells. Oncogene 1995; 11: 1549–1560.
Li Y, Okuno Y, Zhang P, Radomska HS, Chen H, Iwasaki H et al. Regulation of the PU.1 gene by distal elements. Blood 2001; 98: 2958–2965.
Okuno Y, Huang G, Rosenbauer F, Evans EK, Radomska HS, Iwasaki H et al. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol Cell Biol 2005; 25: 2832–2845.
Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet 2004; 36: 624–630.
Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet 2006; 38: 27–37.
Hoogenkamp M, Lichtinger M, Krysinska H, Lancrin C, Clarke D, Williamson A et al. Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood 2009; 114: 299–309.
Yeamans C, Wang D, Paz-Priel I, Torbett BE, Tenen DG, Friedman AD . C/EBPalpha binds and activates the PU.1 distal enhancer to induce monocyte lineage commitment. Blood 2007; 110: 3136–3142.
Ebralidze AK, Guibal FC, Steidl U, Zhang P, Lee S, Bartholdy B et al. PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev 2008; 22: 2085–2092.
Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 2007; 27: 847–859.
Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA 2005; 102: 3627–3632.
Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR et al. Requirement of bic/microRNA-155 for normal immune function. Science 2007; 316: 608–611.
Scott EW, Simon MC, Anastasi J, Singh H . Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 1994; 265: 1573–1577.
Kim HG, de Guzman CG, Swindle CS, Cotta CV, Gartland L, Scott EW et al. The ETS family transcription factor PU.1 is necessary for the maintenance of fetal liver hematopoietic stem cells. Blood 2004; 104: 3894–3900.
Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno S et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 2005; 106: 1590–1600.
Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL . PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med 2005; 201: 1487–1502.
Walsh JC, DeKoter RP, Lee HJ, Smith ED, Lancki DW, Gurish MF et al. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity 2002; 17: 665–676.
McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J 1996; 15: 5647–5658.
DeKoter RP, Walsh JC, Singh H . PU.1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors. EMBO J 1998; 17: 4456–4468.
Olson MC, Scott EW, Hack AA, Su GH, Tenen DG, Singh H et al. PU.1 is not essential for early myeloid gene expression but is required for terminal myeloid differentiation. Immunity 1995; 3: 703–714.
Dakic A, Wu L, Nutt SL . Is PU.1 a dosage-sensitive regulator of haemopoietic lineage commitment and leukaemogenesis? Trends Immunol 2007; 28: 108–114.
Medina KL, Pongubala JM, Reddy KL, Lancki DW, Dekoter R, Kieslinger M et al. Assembling a gene regulatory network for specification of the B cell fate. Dev Cell 2004; 7: 607–617.
Ye M, Ermakova O, Graf T . PU.1 is not strictly required for B cell development and its absence induces a B-2 to B-1 cell switch. J Exp Med 2005; 202: 1411–1422.
Nutt SL, Eberhard D, Horcher M, Rolink AG, Busslinger M . Pax5 determines the identity of B cells from the beginning to the end of B-lymphopoiesis. Int Rev Immunol 2001; 20: 65–82.
Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA . The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell 1990; 61: 113–124.
Hromas R, Orazi A, Neiman RS, Maki R, Van Beveran C, Moore J et al. Hematopoietic lineage- and stage-restricted expression of the ETS oncogene family member PU.1. Blood 1993; 82: 2998–3004.
Nelsen B, Tian G, Erman B, Gregoire J, Maki R, Graves B et al. Regulation of lymphoid-specific immunoglobulin mu heavy chain gene enhancer by ETS-domain proteins. Science 1993; 261: 82–86.
Rekhtman N, Radparvar F, Evans T, Skoultchi AI . Direct interaction of hematopoietic transcription factors PU.1 and GATA- 1: functional antagonism in erythroid cells. Genes Dev 1999; 13: 1398–1411.
Zhang P, Behre G, Pan J, Iwama A, Wara-Aswapati N, Radomska HS et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci USA 1999; 96: 8705–8710.
Nerlov C, Querfurth E, Kulessa H, Graf T . GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood 2000; 95: 2543–2551.
Moreau-Gachelin F, Tavitian A, Tambourin P . Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature 1988; 331: 277–280.
Moreau-Gachelin F, Wendling F, Molina T, Denis N, Titeux M, Grimber G et al. Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Mol Cell Biol 1996; 16: 2453–2463.
Marks PA, Sheffery M, Rifkind RA . Induction of transformed cells to terminal differentiation and the modulation of gene expression. Cancer Res 1987; 47: 659–666.
Marks PA, Rifkind RA . Erythroleukemic differentiation. Annu Rev Biochem 1978; 47: 419–448.
Choe KS, Radparvar F, Matushansky I, Rekhtman N, Han X, Skoultchi AI . Reversal of tumorigenicity and the block to differentiation in erythroleukemia cells by GATA-1. Cancer Res 2003; 63: 6363–6369.
Rao G, Rekhtman N, Cheng G, Krasikov T, Skoultchi AI . Deregulated expression of the PU.1 transcription factor blocks murine erythroleukemia cell terminal differentiation. Oncogene 1997; 14: 123–131.
Zhang P, Zhang X, Iwama A, Yu C, Smith KA, Mueller BU et al. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood 2000; 96: 2641–2648.
Rekhtman N, Choe KS, Matushansky I, Murray S, Stopka T, Skoultchi AI . PU.1 and pRB interact and cooperate to repress GATA-1 and block erythroid differentiation. Mol Cell Biol 2003; 23: 7460–7474.
Stopka T, Amanatullah DF, Papetti M, Skoultchi AI . PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J 2005; 24: 3712–3723.
Behre G, Whitmarsh AJ, Coghlan MP, Hoang T, Carpenter CL, Zhang DE et al. c-Jun is a JNK-independent coactivator of the PU.1 transcription factor. J Biol Chem 1999; 274: 4939–4946.
Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H et al. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell 2007; 1: 416–427.
Du J, Stankiewicz MJ, Liu Y, Xi Q, Schmitz JE, Lekstrom-Himes JA et al. Novel combinatorial interactions of GATA-1, PU.1, and C/EBPepsilon isoforms regulate transcription of the gene encoding eosinophil granule major basic protein. J Biol Chem 2002; 277: 43481–43494.
Cantor AB, Iwasaki H, Arinobu Y, Moran TB, Shigematsu H, Sullivan MR et al. Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. J Exp Med 2008; 205: 611–624.
Sugiyama D, Tanaka M, Kitajima K, Zheng J, Yen H, Murotani T et al. Differential context-dependent effects of friend of GATA-1 (FOG-1) on mast-cell development and differentiation. Blood 2008; 111: 1924–1932.
Tsang AP, Fujiwara Y, Hom DB, Orkin SH . Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev 1998; 12: 1176–1188.
Cantor AB, Orkin SH . Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Semin Cell Dev Biol 2005; 16: 117–128.
Querfurth E, Schuster M, Kulessa H, Crispino JD, Doderlein G, Orkin SH et al. Antagonism between C/EBPbeta and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev 2000; 14: 2515–2525.
Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H et al. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat Immunol 2003; 4: 1029–1036.
Starck J, Cohet N, Gonnet C, Sarrazin S, Doubeikovskaia Z, Doubeikovski A et al. Functional cross-antagonism between transcription factors FLI-1 and EKLF. Mol Cell Biol 2003; 23: 1390–1402.
Bouilloux F, Juban G, Cohet N, Buet D, Guyot B, Vainchenker W et al. EKLF restricts megakaryocytic differentiation at the benefit of erythrocytic differentiation. Blood 2008; 112: 576–584.
Cross MA, Heyworth CM, Murrell AM, Bockamp EO, Dexter TM, Green AR . Expression of lineage restricted transcription factors precedes lineage specific differentiation in a multipotent haemopoietic progenitor cell line. Oncogene 1994; 9: 3013–3016.
Nerlov C, Graf T . PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev 1998; 12: 2403–2412.
DeKoter RP, Singh H . Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 2000; 288: 1439–1441.
Zou GM, Chen JJ, Yoder MC, Wu W, Rowley JD . Knockdown of Pu.1 by small interfering RNA in CD34+ embryoid body cells derived from mouse ES cells turns cell fate determination to pro-B cells. Proc Natl Acad Sci USA 2005; 102: 13236–13241.
Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H . A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity 2009; 31: 576–586.
Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T . Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity 2006; 25: 731–744.
Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER et al. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci USA 2008; 105: 6057–6062.
Hirasawa R, Shimizu R, Takahashi S, Osawa M, Takayanagi S, Kato Y et al. Essential and instructive roles of GATA factors in eosinophil development. J Exp Med 2002; 195: 1379–1386.
Kulessa H, Frampton J, Graf T . GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev 1995; 9: 1250–1262.
Heyworth C, Pearson S, May G, Enver T . Transcription factor-mediated lineage switching reveals plasticity in primary committed progenitor cells. EMBO J 2002; 21: 3770–3781.
Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, Le Beau MM et al. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet 2002; 32: 148–152.
Li Z, Godinho FJ, Klusmann JH, Garriga-Canut M, Yu C, Orkin SH . Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat Genet 2005; 37: 613–619.
Shimizu R, Kobayashi E, Engel JD, Yamamoto M . Induction of hyperproliferative fetal megakaryopoiesis by an N-terminally truncated GATA1 mutant. Genes Cells 2009; 14: 1119–1131.
Steidl U, Steidl C, Ebralidze A, Chapuy B, Han HJ, Will B et al. A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J Clin Invest 2007; 117: 2611–2620.
Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A . High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer 2004; 39: 167–169.
van den Berg A, Kroesen BJ, Kooistra K, de Jong D, Briggs J, Blokzijl T et al. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer 2003; 37: 20–28.
O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med 2008; 205: 585–594.
Shimizu R, Kuroha T, Ohneda O, Pan X, Ohneda K, Takahashi S et al. Leukemogenesis caused by incapacitated GATA-1 function. Mol Cell Biol 2004; 24: 10814–10825.
Mueller BU, Pabst T . C/EBPalpha and the pathophysiology of acute myeloid leukemia. Curr Opin Hematol 2006; 13: 7–14.
Kirstetter P, Schuster MB, Bereshchenko O, Moore S, Dvinge H, Kurz E et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells.[see comment]. Cancer Cell 2008; 13: 299–310.
Miller J, Horner A, Stacy T, Lowrey C, Lian JB, Stein G et al. The core-binding factor beta subunit is required for bone formation and hematopoietic maturation. Nat Genet 2002; 32: 645–649.
Papetti M, Skoultchi AI . Reprogramming leukemia cells to terminal differentiation and growth arrest by RNA interference of PU.1. Mol Cancer Res 2007; 5: 1053–1062.
Blobel GA, Nakajima T, Eckner R, Montminy M, Orkin SH . CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci USA 1998; 95: 2061–2066.
Manabe N, Yamamoto H, Yamada T, Kihara-Negishi F, Hashimoto Y, Mochizuki M et al. Prevention of PU 1-induced growth inhibition and apoptosis but not differentiation block in murine erythroleukemia cells by overexpression of CBP. Int J Oncol 2003; 22: 1345–1350.
Kouzarides T . Chromatin modifications and their function. Cell 2007; 128: 693–705.
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119: 941–953.
Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005; 437: 436–439.
Bhalla KN . Epigenetic and chromatin modifiers as targeted therapy of hematologic malignancies. J Clin Oncol 2005; 23: 3971–3993.
Nome RV, Bratland A, Harman G, Fodstad O, Andersson Y, Ree AH . Cell cycle checkpoint signaling involved in histone deacetylase inhibition and radiation-induced cell death. Mol Cancer Ther 2005; 4: 1231–1238.
Gottlicher M, Minucci S, Zhu P, Kramer OH, Schimpf A, Giavara S et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J 2001; 20: 6969–6978.
Huang ME, Ye YC, Chen SR, Chai JR, Lu JX, Zhoa L et al. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood 1988; 72: 567–572.
Fazi F, Zardo G, Gelmetti V, Travaglini L, Ciolfi A, Di Croce L et al. Heterochromatic gene repression of the retinoic acid pathway in acute myeloid leukemia. Blood 2007; 109: 4432–4440.
Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 2002; 295: 1079–1082.
Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M et al. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature 1998; 391: 815–818.
Lin RJ, Nagy L, Inoue S, Shao W, Miller Jr WH, Evans RM . Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 1998; 391: 811–814.
He LZ, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A et al. Distinct interactions of PML-RARalpha and PLZF-RARalpha with co-repressors determine differential responses to RA in APL. Nat Genet 1998; 18: 126–135.
He LZ, Tribioli C, Rivi R, Peruzzi D, Pelicci PG, Soares V et al. Acute leukemia with promyelocytic features in PML/RARalpha transgenic mice. Proc Natl Acad Sci USA 1997; 94: 5302–5307.
Allenby G, Bocquel MT, Saunders M, Kazmer S, Speck J, Rosenberger M et al. Retinoic acid receptors and retinoid X receptors: interactions with endogenous retinoic acids. Proc Natl Acad Sci USA 1993; 90: 30–34.
Mueller BU, Pabst T, Fos J, Petkovic V, Fey MF, Asou N et al. ATRA resolves the differentiation block in t(15;17) acute myeloid leukemia by restoring PU.1 expression. Blood 2006; 107: 3330–3338.
Acknowledgements
We apologize to the many authors whose work we could not cite owing to space constraints. This work was supported by the grant from Internal Grant Agency of Ministry of Health of the Czech Republic (NR9021-4/2006). Work in P Laslo's laboratory is funded by the Yorkshire Cancer Research and the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Burda, P., Laslo, P. & Stopka, T. The role of PU.1 and GATA-1 transcription factors during normal and leukemogenic hematopoiesis. Leukemia 24, 1249–1257 (2010). https://doi.org/10.1038/leu.2010.104
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2010.104
Keywords
This article is cited by
-
Measuring cell-to-cell expression variability in single-cell RNA-sequencing data: a comparative analysis and applications to B cell aging
Genome Biology (2023)
-
Airway and parenchyma transcriptomics in a house dust mite model of experimental asthma
Respiratory Research (2023)
-
Immortalized erythroid cells as a novel frontier for in vitro blood production: current approaches and potential clinical application
Stem Cell Research & Therapy (2023)
-
The telomere complex and the origin of the cancer stem cell
Biomarker Research (2021)
-
Natural antisense transcripts in the biological hallmarks of cancer: powerful regulators hidden in the dark
Journal of Experimental & Clinical Cancer Research (2020)