Abstract
Complex organismal properties such as longevity can be transmitted across generations by non-genetic factors. Here we demonstrate that deletion of the C. elegans histone H3 lysine 4 dimethyl (H3K4me2) demethylase, spr-5, causes a trans-generational increase in lifespan. We identify a chromatin-modifying network, which regulates this lifespan extension. We further show that this trans-generational lifespan extension is dependent on a hormonal signaling pathway involving the steroid dafachronic acid, an activator of the nuclear receptor DAF-12. These findings suggest that loss of the demethylase SPR-5 causes H3K4me2 mis-regulation and activation of a known lifespan-regulating signaling pathway, leading to trans-generational lifespan extension.
Similar content being viewed by others
Introduction
An increasing number of studies report phenotypes regulated by inheritance of non-Mendelian information from ancestors. For the most part, the molecular mechanisms behind these transgenerational epigenetic effects are still unknown1,2,3. In worms, deletion of SPR-5, the ortholog of the human H3K4me1/2-specific demethylase LSD14, initially do not exhibit phenotypes, however, successive generations lacking this demethylase display increasing infertility concomitant with a global accumulation of DNA methylation of adenines (6mA) and euchromatic histone H3 lysine 4 dimethyl (H3K4me2) and a global decline in heterochromatic H3K9me34,5,6,7. These progressive phenotypes can be reversed by the addition of a single copy of spr-54. Typically, H3K4me2 is associated with gene activation while H3K9me3 is associated with transcriptional repression3. We, and others, previously showed that H3K4me2, H3K9me, and 6mA regulatory proteins regulate the epigenetic memory induced by spr-5 deletion6,7,8. Here we show that lack of SPR-5 not only causes progressive infertility, but also leads to a trans-generational increase in lifespan. The chromatin regulators that suppressed the progressive fertility defect and H3K4me2 and 6mA accumulation also suppressed the trans-generational lifespan extension. Interestingly, we found that the extended trans-generational longevity of spr-5 mutant worms was not simply a byproduct of lowered reproduction but rather a regulated process, involving the known DAF-36/DAF-12 longevity signaling pathway. This DAF-36/DAF-12 signaling pathway did not regulate the inherited H3K4me2 accumulation or reduced egg-laying capacity of spr-5 mutant worms. We further found that addition of dafachronic acid, the steroid which activates DAF-12, was sufficient to extend wild-type (WT) but not late generation spr-5 mutant longevity. Our findings suggest that 6mA, H3K4me2 and H3K9me3 misregulation causes an activation of a known germline to soma signaling pathway which regulates trans-generational effects on longevity.
Results
spr-5 mutant worms display a transgenerational extension of lifespan
Longevity is regulated by genetic and environmental factors9 and has recently been shown, in C. elegans, to be regulated by transgenerational epigenetic inheritance10,11. How epigenetic information is inherited in these instances and how it regulates longevity is still unknown. Because of the role SPR-5 has been shown to play in regulating fertility, the inverse relationship between reproduction and longevity9 and the importance of histone methylation in regulating longevity12, we hypothesized that SPR-5 may also play a role in lifespan regulation. Consistently, one of the top GO enrichment categories for genes regulated by SPR-5 in whole worm expression analyses across generations (generations 1, 13, and 26)4 was 'determination of adult lifespan' (P = 3.4 × 10−4). We found that for the first five generations, two independent spr-5 mutant worm strains, spr-5(by101) and spr-5(by134) (generation 5, G5) had a normal lifespan (Figure 1A and Supplementary information, Table S1). However, after 10 and 20 generations bearing this mutation, spr-5(by101) and spr-5(by134) mutant worms (G10 and G20) displayed extended lifespan by 19%-44% (Figure 1B and 1C, Supplementary information, Tables S1-S5). Both spr-5 alleles are predicted functional null alleles, the by101 allele has a transposon insertion next to the catalytic residue and the by134 allele has a nonsense mutation which eliminates the majority of the enzymatic domain4,5. This trans-generational lifespan extension, similar to the progressive fertility defect and H3K4me2 accumulation, was reverted when worms were backcrossed to provide a single WT copy of spr-5. However, the increase in lifespan was not progressive, i.e., generation 10 spr-5(by101) mutant worms lived as long as generation 20 mutant worms (Figure 1C and Supplementary information, Tables S1 and S2). To determine whether there was a specific generation at which longevity was extended, we examined the lifespan of spr-5(by134) mutant worms at every generation between generation 5 and 10 (Supplementary information, Figure S1 and Table S1). The lifespan extension occurred at generation 7 or generation 8 but was always present by generation 10 (Supplementary information, Figure S1 and Tables S1-S5).
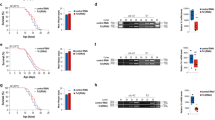
spr-5 mutant worms have extended transgenerational longevity. (A) Early generation (G5) spr-5(by101) mutant worms live as long as wild- type worms (P = 0.3737). Statistics are presented in Supplementary information, Table S1. (B) Late generation (G20) spr-5(by101) mutant worms live longer than wild-type worms (P < 0.0001). Statistics are presented in Supplementary information, Table S1. (C) spr-5(by101) mutant worms do not display a progressive extension in lifespan (bars represent mean ± SEM for two experiments for generation 5, four experiments for generation 10, and 11 experiments for generation 20). Statistics are presented in Supplementary information, Tables S1-S4. (D) spr-5;glp-1 double mutant worms live significantly longer than glp-1 mutant worms similarly to the extension of spr-5 mutant worms lifespan compared with wild-type worms (two-way ANOVA P = 0.3726). glp-1 lifespan extension is compared with WT worms and spr-5;glp-1 lifespan extension is compared with glp-1 mutant worms. Statistics are presented in Supplementary information, Table S2.
To determine whether this trans-generational lifespan extension was simply a byproduct of lowered reproduction or the consequence of alterations of a specific signaling pathway, we examined the lifespan of spr-5(by101) mutant worms whose reproduction was inhibited by either chemical or genetic means. We first treated worms with 5-fluorodeoxyuridine (FUdR), which inhibits proliferation of germline stem cells, the production of intact eggs in adults, and extends longevity13,14. We found FUdR had similar effects on the lifespan of WT and early generation spr-5(by101) mutant worms (G5) (Supplementary information, Figure S2A). However, spr-5(by101) mutant worms after 10 and 20 generations displayed further lifespan extension compared with the WT worms when both were treated with FUdR (Supplementary information, Figure S2B and Table S2), suggesting that this extended longevity was not simply a byproduct of lowered reproduction. To test this hypothesis further, we crossed spr-5(by101) mutant worms with glp-1(e2141ts) mutant worms, which have a somatic germline but fail to develop a meiotic germline and are sterile when maintained at the restrictive temperature after the L1 stage15. spr-5(by101) mutation further extended the long lifespan of glp-1(e2141ts) mutant worms at the restrictive temperature when all strains were carried out to generation 10 (Figure 1D), once again suggesting that the extended trans-generational longevity is not a byproduct of lowered reproduction. Similar results were found when spr-5(by134) mutants were crossed with the temperature sensitive sterile pgl-1(bn101) mutant strain16 (Supplementary information, Table S2). Together, these results suggest that a functional germline is necessary for the transmission of epigenetic information but becomes dispensable for the extended longevity.
Transgenerational longevity of spr-5(by101) mutant worms is dependent on chromatin regulators which control fertility and H3K4me2 and 6mA accumulation
We next investigated whether the extended trans-generational longevity was dependent on the same molecular components that regulate trans-generational fertility defects of spr-5 mutant worms6,7. RNAi inheritance has been implicated in trans-generational epigenetic inheritance in several species17. In C. elegans, exogenous and endogenous RNAi pathways require the argonaute genes rde-1 and ergo-1, respectively18,19, although additional argonautes do exist and could be required for specific RNA inheritance events20. We found that, similar to the progressive fertility defects6, the trans-generationally extended longevity of spr-5 mutant worms was independent of the main exogenous (Supplementary information, Figure S3A) and endogenous (Supplementary information, Figure S3B) RNAi pathways mediated by these argonautes. It remains to be determined whether any of the additional 25 argonautes19, particularly those implicated in heritable RNA21,22,23, could play a role in the trans-generational inheritance of longevity in spr-5(by101) mutant worms. Mutants of some of the untested argonautes display fertility defects and they regulate trans-generational inheritance21 raising the possibility of coordinated regulation of heritable material.
We had previously found that trans-generational fertility defects and H3K4me2 accumulation of spr-5 mutant worms were dependent not only on the euchromatin H3K4 methyltransferases SET-17 and SET-30 but also the H3K9me3/K36me3 demethylase JMJD-2 and the H3K9me binding protein EAP-16. We had also found that the trans-generational fertility defect of spr-5 mutant worms was partially dependent on the potential 6mA methyltransferase DAMT-17. To determine whether these chromatin modifiers additionally controlled the trans-generational longevity extension that we observed in spr-5 mutant worms, we crossed genetic null mutants of these various genes with spr-5(by101) or spr-5(by134) mutant worms and maintained homozygous double mutations with set-17, set-30, jmjd-2, eap-1, or damt-1 for 20 generations. Mutation of set-17, set-30, jmjd-2, or eap-1 was sufficient to completely suppress the extended longevity of spr-5 mutant worms (Figure 2A-2D), as they had suppressed their progressive fertility defects and H3K4me2 accumulations6. Mutation of damt-1 partially suppressed the extended longevity of spr-5 mutant worms (Figure 2E), as it had partially suppressed their progressive fertility defects and H3K4me2 accumulations7.
spr-5 transgenerational longevity is dependent on chromatin regulators which control progressive fertility defect and H3K4me2 and 6mA levels. (A) spr-5(by101) mutant worms live longer than wild-type worms at generation 20 but spr-5;set-17 double mutant worms do not live longer than set-17(n5017) mutant worms (two-way ANOVA P < 0.0001). (B) spr-5(by101) mutation increases the lifespan of wild-type worms but does not increase the lifespan of set-30(gk315) mutant worms after 24 generations (2 way ANOVA, P < 0.0001). (C) spr-5(by101) mutant worms live longer than wild-type worms at generation 20 but spr-5;jmjd-2 double mutant worms do not live longer than jmjd-2(tm2966) mutant worms (two-way ANOVA P = 0.0002). (D) spr-5(by101) mutant worms live longer than wild-type worms at generation 20 but spr-5;eap-1 double mutant worms do not live longer than eap-1(ok3432) mutant worms (2 way ANOVA P < 0.0001). (E) spr-5(by134) mutant worms live longer than wild-type worms at generation 20 and this is partially dependent on damt-1 (two-way ANOVA P = 0.0004). Statistics are presented in Supplementary information, Tables S3 and S4.
Transgenerational longevity but not fertility phenotypes of spr-5(by101) mutant worms is dependent on DAF-12 and DAF-36
Since the trans-generational longevity extension can be decoupled from the progressive fertility defect (Figure 1C and 1D, Supplementary information, Figure S2), we next wanted to determine whether specific signaling pathways, which function downstream of the chromatin-regulating enzymes, regulated the trans-generational longevity. To address this, we examined the spr-5 trans-generationally mis-regulated genes4. Interestingly, the trans-generationally mis-regulated genes in spr-5 mutants analyzed using whole worm extracts4 display significant overlap with daf-12- and daf-16-regulated genes24 (Supplementary information, Figure S4A, hypergeometric probability P = 3.37 × 10−9). All of these daf-12- and daf-16-regulated genes increased expression between generation 13 and generation 1 of spr-54. DAF-12 is a nuclear hormone receptor, which has been shown to be necessary for the longevity extension regulated by a germline to soma longevity signaling pathway25. DAF-12 is activated by the steroid dafachronic acid26, which is synthesized from exogenous cholesterol by a complex biosynthesis process whose first committed step requires the Rieske oxygenase, DAF-3627,28,29. A second major germline to soma signaling pathway that regulates longevity has been reported, which culminates with the transcription factor DAF-16/FOXO9,30.
To address the involvement of these signaling pathways in the extended longevity of spr-5(by101) mutant worms, we first crossed daf-12(m20) with spr-5(by101). We found that late generation spr-5(by101) mutants live longer than their WT counterparts (24.2% longer, P < 0.0001) but the spr-5;daf-12 double mutants did not live significantly longer than daf-12(m20) mutant worms (Figure 3A). Importantly, although removing DAF-12 eliminated the trans-generational extension of lifespan, it had no effect on the global H3K4me2 accumulation (Figure 3B) or the progressive fertility defect (Figure 3C) of spr-5 mutant worms. Taken together, these findings suggest that DAF-12 plays a role in the spr-5-induced lifespan extension but not other progressive phenotypes, suggesting that DAF-12 functions downstream of the chromatin reorganization. Because all of the daf-12-regulated genes increased expression between generation 13 and generation 1 of spr-54, we would hypothesize that prolonged spr-5 loss activates DAF-12.
spr-5(by101) transgenerational longevity is dependent on daf-12 and daf-36. (A) spr-5(by101) mutant worms live longer than wild-type worms at generation 20 but spr-5;daf-12 double mutant worms do not live longer than daf-12(m20) mutant worms (two-way ANOVA P < 0.0001). Statistics are presented in Supplementary information, Table S4. (B) spr-5;daf-12 double mutant worms show similar levels of H3K4me2 as spr-5(by101) mutant worms at generation 20 as assessed by western blots of whole worm lysates of L3 worms. (C) spr-5;daf-12 double mutant worms lay similar numbers of eggs as spr-5(by101) mutant worms at generation 20 (graph is the mean ± SEM of three independent experiments: each experiment consists of average eggs laid for 10 worms of each genotype performed in triplicate). *P < 0.05 by paired t-test; ns, not significant by two-way ANOVA. (D) daf-36 knock down eliminates the spr-5(by101) lifespan extension at generation 15 (2 way ANOVA P < 0.0001). spr-5 EV lifespan extension is compared with WT EV and spr-5 daf-36 lifespan extension is compared with WT daf-36. Statistics are presented in Supplementary information, Table S5. (E) daf-36 knock down does not affect the increased H3K4me2 of spr-5(by101) mutant worms at generation 15 as assessed by western blots of whole worm lysates of L3 worms. (F) daf-36 knock down does not significantly alter the reduced egg laying capacity of generation 15 spr-5(by101) mutant worms. Graph represents the mean ± SEM of 2 independent experiments: each experiment consists of 3 replicates of 10 worms each. ***P < 0.001 by t-test; ns, not significant by two-way ANOVA. (G) Generation 15 spr-5(by101) mutant worms display higher levels of daf-36 mRNA compared with wild-type worms. Graph represents the mean ± SEM of two independent experiments: each experiment consists of 2 biological replicates of 100 worms each. **P < 0.01 by t-test. (H) Dafachronic acid addition extends wild-type worm lifespan (14.5% P < 0.0001) and generation 6 spr-5(by101) mutant worm lifespan (19.6% P = 0.0005). Statistics are presented in Supplementary information, Table S5. (I) Dafachronic acid addition extends wild-type worm lifespan (30.2% P < 0.0001) but does not extend generation 15 spr-5(by101) mutant worm lifespan (3.9% P = 0.2372). Statistics are presented in Supplementary information, Table S5.
To examine this signaling pathway more thoroughly, we knocked down the Rieske oxygenase daf-36, in spr-5(by101) mutant worms. DAF-36 is required for the production of the DAF-12 activating steroid dafachronic acid27,28. Consistently, we found that late generation spr-5(by101) mutant worms treated with daf-36 RNAi eliminated the trans-generational lifespan extension (Figure 3D, two-way ANOVA P < 0.0001). Similar to knockout of daf-12, knockdown of daf-36 also had no effect on the increased H3K4me2 level (Figure 3E) or the progressive fertility decline (Figure 3F). We also found that later generation spr-5 mutant worms displayed elevated mRNA levels of daf-36 (Figure 3G), suggesting that SPR-5 may function to repress daf-36 gene expression. Similar to lifespan extension, elevated daf-36 expression varied from replicate to replicate but was evident by generation 9 (Supplementary information, Figure S4F). Together, these results suggest that the trans-generational longevity extension is separable from the decreased reproduction and requires the DAF-36/DAF-12 signaling pathway.
Since this DAF-36/DAF-12 signaling pathway was required for the extended trans-generational longevity, we next investigated whether all germline to soma longevity signaling pathways were necessary for the extended trans-generational longevity of spr-5 mutant worms. In contrast to DAF-36/DAF-12, our results suggest that DAF-16 was not involved in the extended trans-generational longevity of spr-5 mutant worms. We found that knockdown of daf-16 decreased the longevity of both WT and spr-5(by101) mutant worms by a similar percentage (−13.5% and 16.3%, respectively, two-way ANOVA P = 0.3361) (Supplementary information, Figure S4B). daf-16 knockdown also had no effect on spr-5(by101) egg laying (Supplementary information, Figure S4C). Because results obtained with RNAi are not as conclusive as null alleles31 this experiment leaves open the possibility that the DAF-16 signaling pathway may be involved. To examine this signaling pathway more thoroughly, we also knocked down the intestinal ankyrin-repeat protein kri-1, which is required for DAF-16 nuclear localization and longevity in germline-deficient animals32, in spr-5(by101) mutant worms. We similarly found that late generation spr-5(by101) mutant worms treated with kri-1 RNAi were still long lived compared with WT worms treated with kri-1 RNAi (Supplementary information, Figure S4D, two-way ANOVA P = 0.1831), suggesting that KRI-1 is not involved in spr-5-induced lifespan extension. kri-1 knockdown, like daf-16 knockdown, had no effect on spr-5(by101) egg laying (Supplementary information, Figure S4E). Together, these results suggest that the extended trans-generational longevity, but not other trans-generational phenotypes of spr-5 mutant worms, is dependent on the DAF-36/DAF-12 and not the KRI-1/DAF-16 signaling pathway.
As discussed, the production of dafachronic acid, which is essential for the activation of steroid hormone receptor DAF-12, is dependent on DAF-36. Given that DAF-36 inhibition suppresses the trans-generational longevity phenotype associated with spr-5(by101), we speculate that SPR-5 might regulate lifespan through dafachronic acid via controlling the expression of DAF-36. Consistent with this speculation, the late generation spr-5(by101) worms have elevated levels of daf-36 mRNA (Figure 3G and Supplementary information, Figure S4F). To address this further, we fed WT and spr-5 mutant worms dafachronic acid. We found that dafachronic acid extended the lifespan of WT worms (14.5%, P < 0.0001) and early generation spr-5 mutant worms (19.6%, P = 0.0005) to a similar extent (two-way ANOVA P = 0.4326) (Figure 3H). We found that dafachronic acid extended the lifespan of WT worms (30.2%, P < 0.0001) but not that of generation 15 spr-5(by101) or spr-5(by134) mutant worms (3.9%, P = 0.2372 or 1.8%, P = 0.5661) (Figure 3I). Collectively, these results suggest that increasing dafachronic acid levels extend the lifespan of WT but not late generation spr-5 mutant worms.
Discussion
This study identified a role of SPR-5 in the regulation of trans-generational lifespan. The extended trans-generational longevity is dependent on the same set of chromatin methylation regulators, which control the progressive fertility defect, but is not a byproduct of the reduced fertility. Rather, SPR-5 impacts lifespan by controlling the DAF-12 signaling pathway. Our findings thus lay the framework for a molecular model where the interplay between H3K4 versus H3K9 methylation impacts trans-generational epigenetic inheritance upstream of a specific longevity pathway in C. elegans.
In C. elegans, a reduction in germline stem cell number leads to increased lifespan9, therefore at first glance, it is perhaps not surprising that spr-5(by101), which lay fewer eggs in later generations also live longer. However, egg-laying is not always linked to lifespan14,15. Indeed, we found that spr-5(by101) mutant worms carrying this mutation for 10 generations already reached maximal lifespan extension (Figure 1C). This is different from the trans-generational fertility defect phenotype, which is progressive, i.e., the phenotype continues to worsen until generation 20. Our finding that inhibition of reproduction, either chemically or through genetic removal of the meiotic germline, had no effect on the trans-generational extension of longevity further supports the separation of the fertility and longevity phenotypes. The meiotic germline is required to pass epigenetic information to descendants but after this epigenetic memory has been transmitted it appears dispensable for the extended trans-generational longevity. These experiments do not rule out the possibility that the longevity signal could originate from the somatic gonad which is unaffected by FUdR treatment or the glp-1 or pgl-1 mutations. Since DAF-12 has been shown to be necessary for a germline to soma longevity signaling pathway9,25,30 it could be playing a similar role here. However, this is not necessarily the case since DAF-12 is expressed ubiquitously and the dafachronic synthesis components appear to be expressed throughout the worm in different tissues25. Therefore, the longevity signal could originate from the somatic gonad, from inherited hormones, or from a somatic mis-regulation of chromatin inherited from the parental generation. It will be interesting, in future studies, to determine where this altered longevity cue originates from.
Given that the longevity extension of spr-5 mutants was dependent on a known germline to soma signaling pathway involving DAF-36 and DAF-12 (Figure 3), we speculate that SPR-5, in conjunction with the aforementioned histone methylation regulators, plays a role in the regulation of the DAF-12 target genes. The fact that DAF-36 and DAF-12 manipulation have no effect on reducing global H3K4me2 levels or fertility in spr-5 mutant worms suggests that the chromatin regulation by SPR-5 occurs upstream of the DAF-36/DAF-12 signaling pathway. Consistent with this model, mis-regulated genes in spr-5 mutants display significant overlap with daf-12-regulated genes (Supplementary information, Figure S4A) and spr-5 mutant worms show elevated levels of daf-36 mRNA (Figure 3G). However, it remains to be seen whether SPR-5 directly regulates histone methylation at daf-12-regulated genes. In the future, it will be important to determine if SPR-5 is recruited to these target genes, and if and how SPR-5 deletion modulates the H3K4 methylation regulatory network to promote DAF-12 gene transcription. Finally, does SPR-5 directly inhibit daf-36 gene expression or does it regulate this DAF-36/DAF-12 signaling pathway indirectly through the control of alternative daf-36 regulators? Collectively, our findings suggest that the maintenance of an appropriate chromatin balance is important not only for the generation in which it occurs but also for the longevity of descendants. Our findings provide some of the first mechanistic insight into how epigenetic information from ancestors can influence the longevity of descendants.
It is interesting to note that the previous example of trans-generational epigenetic inheritance of longevity that we identified10 was caused by deletion of an H3K4 trimethylase complex while the example presented here was caused by deletion of an H3K4 mono/dimethyl demethylase. These two examples are very different in that in the first case longevity extension occurs immediately upon deletion of the H3K4 trimethylase complex14 and persists even when WT copies of the H3K4 trimethylase complex are returned and then reverts only after three WT generations to WT longevity10. However, this study finds that initial mutants live as long as WT worms but a longevity extension appears after 7 or 8 generations without the H3K4me2 demethylase. The pathways mediating these two trans-generational longevity phenotypes are distinct: the lifespan extension caused by deletion of the H3K4 trimethylase complex depends on the presence of a functional germline and is independent of daf-1210,14 while the spr-5 deletion induced trans-generational lifespan extension does not require a functional germline (Figure 1D) and is dependent on daf-12 (Figure 3A). In addition, the genes, which are trans-generationally mis-regulated in these two paradigms, do not overlap (data not shown). At the global level, deletion of the H3K4 trimethylase does not affect H3K4me1/me2 levels14 and while H3K4me3 levels are elevated in spr-5 mutant worms they do not become mis-regulated in a trans-generational manner6. H3K4me3 is associated primarily with gene promoters of actively transcribed genes or with genes poised for transcription while H3K4me2 is associated with both promoters and enhancers33. However, both enzymes regulate the methylation of lysine 4 on histone H3. It remains to be determined whether this residue is particularly susceptible to transmitting heritable material or for regulating longevity signaling pathways.
While this provides another example where genetic mis-regulation of a chromatin regulator causes trans-generational lifespan extension, it remains to be seen whether environmental manipulations, which naturally regulate LSD1, can affect trans-generational phenotypes including lifespan. There are increasing examples of how environmental manipulations can have multi- and trans-generational effects on a wide variety of phenotypes1,2,3. There are also correlative reports that environmental alterations can regulate human longevity trans-generationally34. It will be exciting to determine whether these altered trans-generational lifespan phenotypes in people are due to the mis-regulation of H3K4 regulators that we have identified in C. elegans.
Materials and Methods
Lifespan assays
Worm lifespan assays were performed at 20 °C, without 5-fluoro-2′-deoxyuridine (FUdR), as described previously35 unless noted otherwise. When FUdR was used worms were placed on FUdR containing plates at the L4 stage. For each lifespan assay, ∼90 worms per condition were used in three plates to begin the experiment (30 worms per plate). Worms that underwent matricide, exhibited a ruptured vulva, or crawled off the plates, were censored. Statistical analyses of lifespan were performed on Kaplan-Meier survival curves in StatView 5.0.01 by log rank (Mantel-Cox) tests. The values from the Kaplan-Meier curves are included in the Supplementary Tables. For dafachronic acid experiments, (25S)-Δ7-dafachronic acid (Cayman Chemical: 100 μl of 1 mM in EtOH added to 1 liter of media) was added to plate media directly before pouring and worms were kept on dafachronic acid from birth.
Worm strains
The N2 Bristol strain was used as the WT background. The following mutations were used in this study: LG1: spr-5(by101), spr-5(by134); LGII: jmjd-2(tm2966), eap-1(ok3432), set-17(n5017); damt-1(gk961032); LGV: rde-1(ne219), ergo-1(tm1860); and LGX: set-30(gk315), daf-12(m20). Some of these strains have been previously used in4,6,18,19,36. In this paper mutant worms were backcrossed: jmjd-2(tm2966): 2 times, daf-12(m20): 3 times, rde-1(ne219): 3 times, set-17(n5017): 2-7 times, set-30(gk315): 2-8 times, damt-1(gk961032): 5 times, ergo-1(tm1860): 7 times, and eap-1(ok3432): 2-13 times. Generation 1 spr-5 mutant worms were obtained by crossing later generation (generation 10-20) homozygous mutant worms with WT males to obtain P0 heterozygous mutants. Individual P0 heterozygous mutants were picked to plates and allowed to lay generation 1 progeny after which the P0 heterozygous mutant genotype was confirmed by single worm PCR genotyping. Generation 1 progeny were picked to individual plates and allowed to lay progeny. Generation 1 worms were subsequently genotyped by single worm PCR and homozygous mutant worms were perpetuated for subsequent generations. For double mutant crosses WT males were crossed with either spr-5 or second mutant homozygous mutant hermaphrodites. Male progeny from these crosses were then crossed with homozygous mutant or spr-5 hermaphrodites, respectively. Hermaphrodite progeny from these crosses were picked to individual plates and allowed to lay progeny. The parental generation was then genotyped by single worm PCR at the mutant loci used in the male strain. Progeny of heterozygous parental generation worms were picked to individual plates and after laying progeny were genotyped at both mutant loci. Subsequent single or double homozygous mutant progeny were maintained until the appropriate generation and longevity or egg laying assays were performed.
RNA interference
E. coli HT115 (DE3) bacteria containing the vectors of interest were grown at 37 °C and seeded on standard nematode growth medium plates containing ampicillin (100 mg/ml) and isopropylthiogalactoside (IPTG; 0.4 mM). Worms were maintained on dsRNA expressing bacteria starting at generation 2.
Single worm genotyping
Single worms were placed in 5 μl of worm lysis buffer (50 mM KCl, 10 mM Tris pH 8.3, 2.5 mM MgCl2, 0.45% NP40, 0.45% Tween-20, 0.01% gelatin (w/v) and 60 mg/ml proteinase K), and incubated at −80 °C for 1 h, 60 °C for 1 h, and then 95 °C for 15 min. PCR reactions were performed using the following primers: rde-1 F: 5′-TTCATTGAGTTTCCCCACCTACC-3′; rde-1 R: 5′-CTCCTCTGTTTTCATTGGCACC-3′; ergo-1 F: 5′-GCAGGCTTTTAGCGATTTCAAGAC-3′; ergo-1 R: 5′-CGACGGTCAACTCAACTCCATC-3′; set-30 F: 5′-CTCCGTTAGAAGTGGTAGGGGTG-3′; set-30 R: 5′-GAAGTTGCCTCCAAATGCCG-3′; eap-1 F: 5′-TCCATTCAAGTTCCGCAATCC-3′; eap-1 R: 5′-CTCTCCATTAGCATCATTCCCG-3′; spr-5 (by101) F: 5′-AACACGTGCCTCCATGAATATCT-3′; spr-5 (by101) R: 5′-GAACACGTGTGTTCTCCAGCAA-3′; spr-5 (by101) I: 5′-CCTATAGAACTTTCCCACAGTG-3′; spr-5 (by134) F: 5′-CCAATTGTGCTCCAACC-3′; spr-5 (by134) R: 5′-AACTTCGAAGAGCACGGA-3′ (PCR reactions for spr-5(by134) were cut with PsiI to distinguish WT from mutant genotype), set-17 F: 5′-ACCATCTTGCTGTGAAACGAGG-3′; set-17 R: 5′-TGAACGGATTCTTGGCTGGC-3′; jmjd-2 F: 5′-TTTACGCCGCAAAAAAGTGC-3′; jmjd-2 R: 5′-TCTACGATGCTCAAGTGGAAGAGTG-3′; daf-12 F: 5′-GAAAGTTCTGGTGCTTGTGGCTC-3′; daf-12 R: 5′-TGTGGTGACTGCTGATTCCCTG-3′; damt-1 F: 5′-CGGTTATGGAGGAAAGAAGAAGGG-3′; damt-1 R: 5′-TTTTATGGGTATCGGGAACGG-3′; damt-1 I 5′-GCACATCCCAGGAATGAGATTG-3′. PCR reactions were performed according to the manufacturer's protocol (Invitrogen: Platinum PCR supermix) and PCR reactions were resolved on agarose gels.
Antibodies
The H3K4me2 (07-030) was obtained from Millipore. The Histone H3 (ab1791) antibody was obtained from Abcam.
Protein analysis by western blot
Worms were grown synchronously to appropriate stages and washed off plates with M9 buffer. Worms were washed several times in M9 buffer and snap frozen in liquid N2. Sample buffer (2.36% SDS, 9.43% glycerol, 5% β-mercaptoethanol, 0.0945 M Tris-HCl pH 6.8, 0.001% bromophenol blue) was added to worm pellets and they were repeatedly snap frozen in liquid N2. Worm extracts were sonicated three times for 30 s at 15 W (VirSonic 600) and boiled for 2 minutes before being resolved on SDS-PAGE (15%) and transferred to nitrocellulose membranes. The membranes were incubated with primary antibodies (H3K4me2, 1:2 000; H3, 1:2 000) and the primary antibodies were visualized using horseradish peroxidase-conjugated anti-rabbit secondary antibody (Calbiochem 401393) and ECL Plus (Amersham Biosciences).
Worm RNA extraction and reverse transcription followed by Quantitative PCR
RNA was extracted by addition of 1 ml of Trizol (Invitrogen) for 100 μl worm pellets of young adult worms. Six freeze thaw cycles were performed in liquid nitrogen. The RNA extraction was performed according to the Trizol protocol. The expression of target genes was determined by reverse transcription of 1 μg total RNA with the Superscript III kit (Invitrogen) followed by quantitative PCR analysis on a Roche Lightcycler 480 II with SYBR Green I Master (Roche) with the following primers: pan-actin F: 5′-TCGGTATGGGACAGAAGGAC-3′; pan-actin R: 5′-CATCCCAGTTGGTGACGATA-3′; daf-36 F: 5′-GGAGGATAGACATTGTGGAGTTATGC-3′; daf-36 R: 5′-CGGGAGTTACTGTTTGAAATACGAC-3′. The results were expressed as 2(−(gene of interest number of cycles − actin number of cycles)). Control PCR reactions were also performed on total RNA that had not been reverse-transcribed to test for the presence of genomic DNA in the RNA preparation.
Author Contributions
ELG and YS conceived and planned the study and wrote the paper. ELG performed the experiments. BB, CL, and AA analyzed steroid concentrations.
Competing Financial Interests
YS is a co-founder of Constellation Pharmaceuticals, Inc and a member of its scientific advisory board. The remaining authors declare no conflict of interest.
References
Youngson NA, Whitelaw E . Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet 2008; 9:233–257.
Daxinger L, Whitelaw E . Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet 2012; 13:153–162.
Greer EL, Shi Y . Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012; 13:343–357.
Katz DJ, Edwards TM, Reinke V, Kelly WG . A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 2009; 137:308–320.
Nottke AC, Beese-Sims SE, Pantalena LF, Reinke V, Shi Y, Colaiácovo MP . SPR-5 is a histone H3K4 demethylase with a role in meiotic double-strand break repair. Proc Natl Acad Sci USA 2011; 108:12805–12810.
Greer EL, Beese-Sims SE, Brookes E, et al. A histone methylation network regulates transgenerational epigenetic memory in C. elegans. Cell Rep 2014; 7:123–126.
Greer EL, Blanco MA, Gu L, et al. DNA methylation on N(6)-Adenine in C. elegans. Cell 2015; 161:868–878.
Kerr SC, Ruppersburg CC, Francis JW, Katz DJ . SPR-5 and MET-2 function cooperatively to reestablish an epigenetic ground state during passage through the germ line. Proc Natl Acad Sci USA 2014; 111:9509–9514.
Kenyon CJ . The genetics of ageing. Nature 2010; 464:504–512.
Greer EL, Maures TJ, Ucar D, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 2011; 479:365–371.
Rechavi O, Houri-Ze'evi L, Anava S, et al. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell 2014; 158:277–287.
Benayoun BA, Pollina EA, Brunet A . Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol, 2015; 16:593–610.
Mitchell DH, Stiles JW, Santelli J, Sanadi DR . Synchronous growth and aging of Caenorhabditis elegans in the presence of fluorodeoxyuridine. J Geront 1979; 34:28–36.
Greer EL, Maures TJ, Hauswirth AG, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 2010; 466:383–387.
Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C . Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science 2002; 295:502–505.
Kawasaki I, Shim YH, Kirchner J, Kaminker J, Wood WB, Strome S . PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 1998; 94:635–645.
Moazed D . Mechanisms for the inheritance of chromatin States. Cell 2011; 146:510–518.
Grishok A, Tabara H, Mello CC . Genetic requirements for inheritance of RNAi in C. elegans. Science 2000; 287:2494–2497.
Yigit E, Batista PJ, Bei Y, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 2006; 127:747–757.
Conine CC, Moresco JJ, Gu W, et al. Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell 2013; 155:1532–1544.
Buckley BA, Burkhart KB, Gu SG, et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 2012; 489:447–451.
Ashe A, Sapetschnig A, Weick EM, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 2012; 150:88–99.
Simon M, Sarkies P, Ikegami K, et al. Reduced insulin/IGF-1 signaling restores germ cell immortality to Caenorhabditis elegans Piwi mutants. Cell Rep 2014; 7:762–773.
McCormick M, Chen K, Ramaswamy P, Kenyon C . New genes that extend Caenorhabditis elegans' lifespan in response to reproductive signals. Aging Cell 2012; 11:192–202.
Antebi A . Steroid regulation of C. elegans diapause, developmental timing, and longevity. Curr Top Dev Biol 2013; 105:181–212.
Motola DL, Cummins CL, Rottiers V, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell 2006; 124:1209–1223.
Wollam J, Magomedova L, Magner DB, et al. The Rieske oxygenase DAF-36 functions as a cholesterol 7-desaturase in steroidogenic pathways governing longevity. Aging Cell 2011; 10:879–884.
Rottiers V, Motola DL, Gerisch B, et al. Hormonal control of C. elegans dauer formation and lifespan by a Rieske-like oxygenase. Dev Cell 2006; 10:473–482.
Yoshiyama-Yanagawa T, Enya S, Shimada-Niwa Y, et al. The conserved Rieske oxygenase DAF-36/Neverland is a novel cholesterol-metabolizing enzyme. J Biol Chem 2011; 286:25756–25762.
Mukhopadhyay A, Tissenbaum HA . Reproduction and longevity: secrets revealed by C. elegans. Trends Cell Biol 2007; 17:65–71.
Gems D, Pletcher S, Partridge L . Interpreting interactions between treatments that slow aging. Aging Cell 2002; 1:1–9.
Berman JR, Kenyon C . Germ-cell loss extends C. elegans lifespan through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell 2006; 124:1055–1068.
Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007; 39:311–318.
Pembrey ME, Bygren LO, Kaati G, et al. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet 2006; 14:159–166.
Greer EL, Dowlatshahi D, Banko MR, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol 2007; 17:1646–1656.
Whetstine JR, Nottke A, Lan F, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 2006; 125:467–481.
Acknowledgements
We thank Lieberman J, Blackwell TK, and members of the Shi lab for discussions and critical reading of the manuscript. We thank Liefke R for bioinformatic support. We thank Stiernagle T and the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), Mitani S and the National Bioresource Project for the Experimental Animal 'Nematode C. elegans', and Moerman D laboratories for C. elegans strains. ELG was supported by a Helen Hay Whitney post-doctoral fellowship and by a National Institute on Aging of the National Institute of Health (NIH) grant (AG043550). BB, CL, and AA were supported by the Max Planck Gesselschaft. This work was supported by a grant from the NIH to YS (GM058012) and by an Ellison Foundation Senior Scholar Award to YS. YS is an American Cancer Society Research Professor.
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Table S1
Spr-5(by101) and spr-5(by134) mutant worms have a transgenerational extended longevity (PDF 52 kb)
Supplementary information, Table S2
spr-5 mutant worms lifespan doesn't require egg laying and depends on eap-1 (PDF 56 kb)
Supplementary information, Table S3
Spr-5(by101) mutant worms have a transgenerational extended longevity which is dependent on eap-1, set-17, and set-30 but independent of ergo-1 and rde-1 (PDF 57 kb)
Supplementary information, Table S4
Spr-5(by101) mutant worms have a transgenerational extended longevity which is dependent on eap-1, jmjd-2, damt-1 and daf-12 (PDF 56 kb)
Supplementary information, Table S5
spr-5(by101) mutant worms' transgenerational extended longevity is dependent on daf-36 but independent of daf-16 and kri-1 and dafachronic acid extends the lifespan of wildtype but not late generation spr-5 mutant worms (PDF 58 kb)
Supplementary information, Figure S1
spr-5 mutants display extended transgeneraitonal longevity by generation 7 or 8 (PDF 85 kb)
Supplementary information, Figure S2
Extended transgenerational longevity of spr-5 mutants is independent of reduced egg laying (PDF 415 kb)
Supplementary information, Figure S3
Extended transgenerational longevity of spr-5 mutants is independent of rde-1 and ergo-1 (PDF 440 kb)
Supplementary information, Figure S4
Extended transgenerational longevity and reduced fertility of spr-5 mutant worms is independent of daf-16 and kri-1 (PDF 533 kb)
Rights and permissions
About this article
Cite this article
Greer, E., Becker, B., Latza, C. et al. Mutation of C. elegans demethylase spr-5 extends transgenerational longevity. Cell Res 26, 229–238 (2016). https://doi.org/10.1038/cr.2015.148
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2015.148
Keywords
This article is cited by
-
Aging in the Drosophila ovary: contrasting changes in the expression of the piRNA machinery and mitochondria but no global release of transposable elements
BMC Genomics (2019)
-
Regulation of lifespan by neural excitation and REST
Nature (2019)
-
Non-genomic transmission of longevity between generations: potential mechanisms and evidence across species
Epigenetics & Chromatin (2017)