Abstract
Linker of the nucleoskeleton and the cytoskeleton (LINC) complexes are composed of SUN and KASH domain-containing proteins and bridge the inner and outer membranes of the nuclear envelope. LINC complexes play critical roles in nuclear positioning, cell polarization and cellular stiffness. Previously, we reported the homotrimeric structure of human SUN2. We have now determined the crystal structure of the human SUN2-KASH complex. In the complex structure, the SUN domain homotrimer binds to three independent “hook”-like KASH peptides. The overall conformation of the SUN domain in the complex closely resembles the SUN domain in its apo state. A major conformational change involves the AA'-loop of KASH-bound SUN domain, which rearranges to form a mini β-sheet that interacts with the KASH peptide. The PPPT motif of the KASH domain fits tightly into a hydrophobic pocket on the homotrimeric interface of the SUN domain, which we termed the BI-pocket. Moreover, two adjacent protomers of the SUN domain homotrimer sandwich the KASH domain by hydrophobic interaction and hydrogen bonding. Mutations of these binding sites disrupt or reduce the association between the SUN and KASH domains in vitro. In addition, transfection of wild-type, but not mutant, SUN2 promotes cell migration in Ovcar-3 cells. These results provide a structural model of the LINC complex, which is essential for additional study of the physical and functional coupling between the cytoplasm and the nucleoplasm.
Similar content being viewed by others
Introduction
In eukaryotic cells, the nuclear envelope (NE) separates the nucleoplasm from the cytoplasm, which is important for maintaining the integrity of genetic material and the complex regulation of gene expression and function. The NE consists of two lipid bilayers, the inner and the outer nuclear membrane (INM and ONM), which are connected at nuclear pores. The ONM is contiguous with the endoplasmic reticulum membrane, and the INM adheres to the nuclear lamina matrix. Only the nuclear pore complex has been well studied, and only minimal knowledge about the remaining NE exists. Only recently have molecular characterization and studies of the functional importance of the nuclear double membrane architecture emerged. For example, the linker of the nucleoskeleton and the cytoskeleton (LINC) complex has been identified as a molecular bridge across the NE for the transduction of signaling and mechanical force1,2. LINC complexes primarily consist of proteins containing a SUN (Sad1 and UNC84) domain or a KASH (Klarsicht, ANC-1 and Syne homology) domain, which function as a part of the mechano/signaling cascade from the cellular periphery to the genome.
As core components of the LINC complexes, both the SUN and KASH domains are evolutionarily conserved, supporting a fundamental and conserved role of LINC complexes. Indeed, mounting evidence indicates that LINC complexes function in multiple biological processes, including nuclear migration and repositioning, meiotic telomere tethering and chromatin organization, centrosome migration and attachment, nucleokinesis and cellular rigidity homeostasis1,2,3. Recently, LINC complexes have been implicated in the regulation of apoptosis4,5, the maturation and survival of germlines6, and in the pathology of human diseases linked to mutations of A-type lamins, such as laminopathies and Emery-Dreifuss muscular dystrophy7,8,9.
During a study of centrosome-related proteins that were affected by erbB transformation, we cloned and identified the SUN domain protein SUN210. Human SUN2 is a ubiquitously expressed type II transmembrane protein that localizes at the INM. A transmembrane domain separates the N-terminal nucleoplasmic region, which interacts with lamina and/or chromatin binding proteins, from the C-terminal luminal region, which protrudes into the perinuclear space. The luminal region of SUN2 contains two coiled-coil domains and a conserved SUN domain at the C-terminus11 (Figure 1A). According to our recent structural study of SUN2, the conserved SUN domain is essential and sufficient for KASH binding12. Endogenous SUN2 is localized to the NE and is partly dependent on lamin A, but not lamin C, for NE retention13. SUN2 is targeted to the INM through classical nuclear localization signal motifs at the N-terminus and the perinuclear SUN domain at the C-terminus14. SUN2 appears to associate with the centrosome in a cell cycle-dependent manner. Previously, we showed that SUN2 may form homo-oligomers, which is regulated by its two coiled-coil regions and the SUN domain10. SUN2 can also form hetero-oligomers with SUN1, another SUN domain protein that is associated with the nuclear pore complex10,15. Both SUN1 and SUN2 are widely expressed KASH-binding partners and may have partially redundant functions. Double knockout of SUN1 and SUN2 is lethal in mice, similar to the disruption of KASH domain-containing proteins, highlighting the functional importance of the LINC complex16.
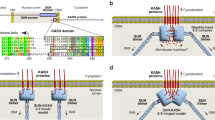
Overall structure of the SUN-KASH complex. (A) A schematic illustration of the domain organization for human SUN2 and Nesprin 2G. (B) The top and side views of the SUN domain homotrimer shown in cartoon representation. The three protomers of the SUN homotrimer are colored in cyan, light blue and pale green. (C) The top and side views of the SUN-KASH complex. The three KASH domain peptides are colored in yellow, magenta and red and bind to three separate trimeric interfaces of the SUN domain homotrimer.
Upon formation of the LINC complex, the INM proteins with SUN domains recruit the proteins with a KASH domain to the OMN via interactions between their SUN and KASH surfaces17. Nesprins are representative KASH domain proteins that contain a single transmembrane anchor at the C-terminus for ONM localization. The N-terminal domains of KASH proteins are highly diversified and are associated with the cytoskeleton by connecting to actin filaments, microtubules and intermediate filaments. The conserved C-terminal KASH domain consists of approximately 35 amino acids that are exposed to the luminal space of the nuclear double membrane for interaction with the SUN domain of INM proteins (Figure 1A). In the lumen of the NE, the SUN domain interacts with the KASH domain, ensuring that KASH domain proteins are targeted to the ONM. During actin-dependent nuclear movement, the LINC complex components Nesprin 2G and SUN2 were found to provide a direct linkage between the nucleus and the actin network18,19. Recently, mutations of KASH domain proteins have been linked to Emery-Dreifuss muscular dystrophy20,21.
The NE-spanning LINC complexes that are formed through interactions between the SUN domain and the KASH domain exemplify a physical linkage that may affect many aspects of cellular functions. However, the assembly of the SUN-KASH core complex has not been fully defined at the atomic level, hindering further exploration of its functions. Based on our previous crystallographic study of the SUN domain12, we performed structural, biochemical and cell-based studies of the SUN-KASH complex. We observed a 3:3 ratio in the hexameric SUN-KASH complex in which the SUN domain trimer binds three separate KASH peptides. Our results also indicate that the two regions corresponding to the first and second coiled-coil motifs preceding the SUN domain may function as activation and inhibitory domains, respectively, for its KASH-binding activity.
Results
Overall structure of the SUN-KASH complex
We previously reported the crystal structure of the SUN domain of human SUN2 as a homotrimer and mapped the essential domains required for SUN-KASH association12. We demonstrated that a synthesized peptide (termed the KASH peptide), corresponding to the conserved C-terminus of the human Nesprin 2G KASH domain, can bind to the SUN domain of human SUN2 and that disruption of the SUN domain homotrimer abolished the binding of the KASH peptide. To further understand the structural basis of the LINC complex assembly and its functional mechanism at an atomic level, we crystallized the SUN2 SUN domain (G522-H717) in complex with the conserved KASH domain peptide (SFYPMLRYTNGPPPT). The crystal structure of the SUN-KASH complex was determined to 3.05 Å resolution by molecular replacement using the SUN domain monomer as a search model (PDB code 3UNP).
In the SUN-KASH complex structure, the SUN domain forms a cloverleaf-like homotrimer with an overall conformation that is highly similar to that observed for the apo-SUN domain homotrimer (Figure 1B). Briefly, the SUN domain monomer is composed of an N-terminal stem-like α-helix and a C-terminal leaf-like β-sandwich. Three helices from the individual protomers bundle together to form the stem of the SUN domain homotrimer. The β-sandwich subdomains pack against one another to form the threefold symmetrical lobes of the trimer.
Consistent with our previous structural and biochemical studies, the SUN domain homotrimer binds three separate KASH peptides, forming a 3:3 hexameric heterocomplex (Figure 1C). Notably, the three individual KASH peptides bind to three separate sites, which are symmetrically distributed close to the trimeric interface of the SUN domain homotrimer. Overall, the KASH domain is shaped like a “hook” that is embedded in the SUN domain trimer. The AA'-loop of one protomer and the β-sandwich of the neighboring protomer clamp the KASH domain, whose kinked C-terminus extends into a hydrophobic pocket on the SUN domain surface near β-strands B and I (termed as the BI-pocket). Two neighboring SUN domain protomers interact with the sandwiched KASH peptide, burying 834.6 Å2 and 426 Å2 solvent accessible surface areas, respectively.
Comparison of the SUN-KASH complex with the SUN domain
The overall structures of the complexed and apo-SUN domain are very similar, with an RMSD value of 0.616 Å for 195 Cα atoms of the overlapped SUN domain structures. However, the structural comparison does reveal a striking conformational change in the AA'-loop of the SUN domain between the apo-state and the complexed-state (Figure 2A). In the apo-SUN domain, a major part of the AA'-loop, especially its tip, is poorly defined or disordered, which indicates its flexibility in the absence of ligand (Figure 2B). When in complex with the KASH domain, the AA'-loop of the SUN domain is a well-defined mini β-sheet consisting of two anti-parallel β-strands on the tip (Figure 2C). It is apparent that the KASH domain, which also folds into a β-strand, may induce the conformational change of the SUN domain AA'-loop and further stabilize the rearranged conformation by forming a three-stranded intermolecular β-sheet.
Comparison of the SUN-KASH complex with the apo-SUN domain. (A) KASH-binding induced a conformational change of the AA'-loop of the SUN domain. The AA'-loop is poorly defined and partially disordered in the apo-SUN domain structure. In the SUN-KASH complex, the AA'-loop folds into a two-stranded anti-parallel mini β-sheet, and together with the bound KASH strand, forms an intermolecular β-sheet. (B) A surface presentation of the apo-SUN domain. The AA'-loop is partially disordered. (C) A surface presentation of the SUN domain in complex with the KASH peptide.
Interface between the SUN domain trimer and the KASH peptides
The KASH domain directly interacts with two adjacent protomers of the SUN domain homotrimer (Figure 3A), with three major contact sites at the SUN-KASH interface. At the first site, the C-terminal PPPT motif of the KASH domain fits into the hydrophobic BI-pocket on the SUN domain surface with perfect complementarity (Figure 3B). The KASH domain forms hydrophobic interactions with residues Ala603 from the B strand, Tyr703 from the I strand and Tyr707 from the loop between the I and J strands. Additionally, residue Thr6885 of KASH domain forms hydrogen bonds with residues Ser641 from the DE-loop and Tyr707 from the IJ-loop. Residue Tyr703 from the I strand of the SUN domain also forms contacts with the main chain atoms of the KASH peptide. Residue Cys601 from the B strand forms an intramolecular disulfide bond with Cys705, which may promote KASH binding.
Interface between the SUN domain trimer and the KASH peptides. (A) The SUN domain is shown as a surface model, and the KASH domain is shown in a cartoon and stick model for clarity. The KASH domain is colored yellow. The interface includes three interaction sites (see text). (B) A zoom-in picture of site 1 of the SUN-KASH interface. The PPPT motif embeds in a hydrophobic BI-pocket in the SUN domain. The SUN domains are colored in cyan and light blue, and the KASH domain is colored in yellow. (C) A zoom-in picture of sites 2 and 3 of the SUN-KASH interface. Two protomers of the SUN domain homotrimer sandwich the β-strand of the KASH domain. The tip of the AA'-loop from one protomer and the base of the AA'-loop from the adjacent protomer clamp the KASH domain.
In addition to the C-terminal hydrophobic docking, the remainder of the KASH peptide is sandwiched by the extended tip of the AA'-loop from one protomer of the SUN domain trimer and the A' helix near the base of the AA'-loop from another protomer to form a distorted intermolecular β-sheet. This sandwiching interaction can be divided into the second and third sites. At the second site, residues Arg6877, Thr6879, Asn6880 and Gly6881 are exposed to the solvent, and residue Thr6879 and the main chain of the KASH domain form hydrogen bonding with residues Thr569 and Tyr583 in the AA'-loop (Figure 3C).
At the third site, residues Met6875 of the KASH domain and Ile559 of the SUN domain form a hydrophobic interaction. Residue Tyr6873 and the main chain of the KASH domain form hydrogen bonds with residues His584 and Arg589 of the SUN domain. Additionally, the terminal Phe6872 of the KASH domain makes contact with the hydrophobic Trp582 of the SUN domain (Figure 3C).
Structure-based mutational analyses of the SUN-KASH complex assembly
To analyze the contribution of individual amino acids to complex formation, we made structure-based truncations and point mutations on both the SUN and KASH domains. For the GST pull-down and Octet experiments, we used purified recombinant proteins. All mutant proteins of the SUN domain were first subjected to analysis by gel filtration to confirm that their global folding and oligomerization were not affected by the mutations introduced in this work. To mimic the KASH domain in vitro, we fused the C-terminal part of Nesprin 2G (Ser6856-Thr6885) to GST (GST-KASH) and tested its interaction with the wild-type (WT) human SUN2 SUN domain. As shown in Figure 4A, the WT SUN domain could robustly bind to GST-KASH, indicating that our in vitro assay systems were legitimate for characterization of the SUN-KASH complex.
In vitro analyses of the SUN-KASH interactions. (A) WT and mutant SUN domain proteins were subjected to GST pull-down by GST-KASH and GST-KASHΔ fusion proteins (See details in Materials and Methods). (B) Kinetic analysis of the affinity of WT or mutant SUN domains and KASH domain by Octet Red 96. (C) The SUN2 luminal regions containing different coiled-coil motifs were incubated with Glutathione-Sepharose resin loaded with GST-KASH. CC1-CC2-SUN was incubated with Glutathione-Sepharose resin loaded with GST as control. (D) Kinetic analysis of the affinity of SUN2 luminal region containing different coiled-coil motifs and KASH domain. The bands of various SUN2 proteins were labeled by asterisk in A and C.
Our current structure revealed intimate hydrophobic docking of four C-terminal residues of the KASH domain (PPPT motif) into the BI-pocket of the SUN domain, which is consistent with earlier studies showing that the conserved PPPT motif of the KASH domain was involved in association with the SUN proteins22,23,24. Based on structural analysis, we tested the contribution of this interaction by GST pull-down assay using a KASH peptide with a deletion of C-terminal PPPT motif (GST-KASHΔ). Our result clearly showed that removal of the PPPT motif abolished KASH association with the SUN domain, indicating an essential role of this motif in LINC complex formation (Figure 4A). A previous study by Stewart-Hutchinson et al.22 indicated that additional residues added at the C-terminus of the KASH domain prevented KASH from binding with SUN proteins. This observation can be explained by the structural feature of the BI-pocket of the SUN domain, which can only sterically accommodate a ligand of the size of the PPPT motif.
To assess the importance of the AA'-loop in SUN-KASH binding, we deleted the extended tip of the AA'-loop (del AA', Y567-H584) for interaction analysis. As shown by both GST pull-down and Octet analysis, the WT SUN domain binds to GST-KASH with a Kd of 45.6 nM, whereas the del AA' mutant SUN domain could only weakly bind to GST-KASH. The affinity of the del AA'-SUN domain and the KASH domain could not be determined by Octet assay (Figure 4A and 4B), suggesting that the formation of the intermolecular β-sheet between the AA'-loop and the KASH domain can greatly stabilize the anchoring of the PPPT motif by the BI-pocket. Of note, our previous work showed that deletion of the SUN domain stem region disrupted the homotrimer and completely abolished its KASH-binding ability12. This observation, together with the SUN-KASH structure, indicates that the sandwiching of the KASH domain by two protomers of the SUN domain homotrimer is critical for complex assembly.
Furthermore, to examine the residues that are critical for KASH binding, we mutated several conserved amino acids of the SUN domain that are located near the hydrophobic pocket of the first interaction site. Mutations A603E from the B strand, Y703E from the I strand and S641E from the DE-loop greatly impaired the SUN-KASH interaction. Moreover, a three-point mutation of the SUN domain (A604E/S641E/Y703E) completely abolished KASH binding (Figure 4A). Mutation Y707A from the IJ-loop also disrupted the interaction between the SUN domain trimer and the KASH domain (Figure 4A). Disturbing the SUN domain BI-pocket by introduction of mutations, including A603E, Y703E, S641E and Y707A, impaired or abolished the GST-KASH binding activity, highlighting the importance of this docking site. Consistent with our previous work12, mutation of the SUN domain surface residue G609D also disrupted the SUN-KASH assembly, which was most likely an indirect outcome caused by local conformational changes (Figure 4A).
We investigated the potential regulatory effects of the SUN2 region outside of the SUN domain on KASH binding. The two coiled-coil motifs within the luminal domain of human SUN2 have been predicated to be most likely dimeric (CC1) and trimeric (CC2)12. We tested the interaction between the KASH domain and the SUN2 luminal region containing different coiled-coil motifs by GST pull-down and Octet assays. As expected, the SUN2 fragment containing the two coiled-coil motifs (CC1 and CC2) and the intact SUN domain (CC1-CC2-SUN) could effectively bind to the KASH domain, but not the GST control (Figure 4C). Moreover, deletion of the AA'-loop of the CC1-CC2-SUN construct attenuated its KASH-binding ability. However, a fragment containing the second coiled-coil motif (CC2) and the intact SUN domain (CC2-SUN) was incapable of binding to the KASH domain (Figure 4C), which is consistent with a previous report that removal of the CC1 of SUN1 abrogated its interaction with Nesprin 2G1. Octet analysis of the affinity of the SUN2 luminal region containing different coiled-coil motifs and the KASH domain was compatible with the GST pull-down assay (Figure 4D). Given that homo-trimerization of the SUN domain is required for the sandwich docking of the KASH domain on the trimeric interface, it is possible that SUN2 region containing the coiled-coil motifs regulates SUN-KASH (de)coupling through the modulation of SUN domain oligomerization.
The SUN-KASH complex is involved in cell migration
Previous studies have shown that knockout of Nesprin 2G impaired cell migration and proliferation, likely by disrupting LINC complex assembly25. To gain functional insights into the SUN-KASH complex, we investigated the cell migration effect based on our atomic and biochemical observations of the SUN-KASH interactions. We transfected Ovcar-3 cells with full-length WT or mutant human SUN2 and used pcDNA 3.1 and Myc-FAK as a negative and positive control, respectively. After 24 h, cell migration was evaluated using a transwell assay. It has been well established that focal adhesion kinase (FAK) promotes cell migration through the growth-factor-receptor (GFR) and integrin signaling pathways26,27. As shown in Figure 5, human SUN2 promoted cell migration, similar to the positive control FAK; the percentage of cells that migrated through the membrane was 2-fold higher than the control. However, various mutants with mutations in the SUN domain of SUN2, including del AA', A603E, G609D, S641E, Y703E and Y707A, had no promoting effect on cell migration, as the percentages of cells that migrated through the membrane were similar to cells transfected with pcDNA 3.1 (Figure 5). These results are consistent with those from the GST pull-down and kinetics analyses, and we propose that disruption of the SUN-KASH complex assembly impairs the function of SUN2 in cell migration.
The SUN-KASH complex is involved in cell migration. (A-I) 1 × 105 Ovcar-3 cells transfected with pcDNA3.1, Myc-FAK, Flag-tagged WT or mutant SUN2 and RPMI 1640 supplemented with 10% fetal bovine serum were added to the top and the bottom chamber, respectively. 24 h later, the cells that migrated to the bottom surface of the inserts were detected with staining and light microscopy. (J) Quantitative analysis of the effects of WT SUN2 and various SUN2 mutants on cell migration in Ovcar-3 cells.
Discussion
Our previous and current work reveals a homotrimeric structure of the SUN domain, which resembles a clover leaf12. Here, we reveal the LINC complex as a 3:3 hexameric SUN-KASH assembly and identify the BI-pocket, the AA'-loop and the homotrimeric interface of the SUN domain as key structural determinants for KASH binding. As the bound KASH domain is essentially sandwiched by two adjacent protomers of the SUN domain trimer, the trimeric conformation of the SUN domain is a prerequisite for LINC complex formation. Indeed, disruption of the SUN domain homotrimer completely abolished KASH binding12. Deletion of the AA'-loop substantially reduced the stability of the SUN-KASH complex, again reflecting the sandwich nature of KASH binding (Figure 4). Individual mutations that modify the hydrophobic BI-pocket of SUN domain reduced or abolished KASH binding, corroborating the importance of this pocket as the docking site for the KASH domain.
The KASH domain peptide in the complex structure folds into a short β-strand followed by a kinked linker and a PPPT motif (Figure 3). The β-strand is sandwiched by the SUN domain protomers, stabilizing the SUN-KASH complex by forming an intermolecular β-sheet with the AA'-loop of the SUN domain. We found that removal of the PPPT motif at the C-terminus of the human Nesprin 2G KASH domain disrupted its binding to human SUN2. This terminal PPPT motif functions as a primary anchor by tightly fitting into the BI-pocket of the SUN domain. The addition of amino acids at the C-terminus of the PPPT anchor leads to steric hindrance of PPPT docking into the BI-pocket. These observations are consistent with a previous report showing that an extension of the C-terminal KASH domain sequence prevented the binding to the SUN domain22,23,24.
By transducing mechanical force across the NE, the LINC complex has been implicated in many cellular events, such as nuclear migration and positioning16,28. Interestingly, the KASH protein Nesprin 2G has also been shown to positively regulate cell migration25. Other studies suggest that SUN-KASH complexes are necessary for cell migration in C. elegans and Arabidopsis29,30,31. We found that over-expression of WT SUN2 promotes cell migration on a scale similar to FAK. However, mutant forms of SUN2 that disrupted the SUN-KASH interaction did not enhance cell migration. Moreover, a mutation of the SUN protein UNC84, corresponding to G609D of human SUN2, leads to defects in cell migration in C. elegans32. We found that the G609D mutation greatly compromised the SUN-KASH assembly in vitro (Figure 4), and the corresponding mutant SUN2 did not promote cell migration (Figure 5). The residue G609 is located on the surface of the SUN domain β-sandwich, which would not directly affect KASH binding. As the G609 residue is in the vicinity of the AA'-loop and the BI-pocket, we suggest that the G609D mutation may disturb the conformation of these two structural determinants, causing disassembly of the SUN-KASH complex. Our results correlate the assembly of the LINC complex with cell functions and suggest that the LINC complex may play a positive role in cell migration. We suggest that WT SUN2 may facilitate the coordination between the actin network and the nucleus through enhanced transduction of mechanical force, while mutant forms of SUN2 fail to do so. SUN-KASH complexes have been implicated in many fundamental biological processes. It is now possible to reinvestigate these cellular aspects using mutational information derived from our structural analysis. Given the conservation of the LINC complex from plants to humans, it is likely that the SUN-KASH bridges may universally relay the mechanical force across the NE to coordinate multiple cellular events.
Previously, we demonstrated that deletion of the stem-like α-helical subdomain of the SUN domain completely disrupted its trimerization, indicating that the leaf-like β-sandwich subdomain is not sufficient for the formation of a stable SUN domain homotrimer12. Given that homotrimerization of the SUN domain is a prerequisite for KASH binding, the α-helical subdomain could serve as a regulatory handle for LINC complex assembly and/or decoupling. In fact, we have repeatedly observed that both the SUN domain homotrimer alone and the luminal region of SUN2 containing two coiled-coils and the SUN domain (CC1-CC2-SUN) are sufficient for KASH binding (Figure 4). However, a truncated form of SUN2 that only contains the second coiled-coil and the SUN domain (CC2-SUN) was not able to bind the KASH peptide, consistent with previous studies1,10,22,24. Of note, we previously found that the truncated form containing only CC2 and the SUN domain also showed reduced oligomerization10. We propose that the region corresponding to the second coiled coil (CC2) functions as an inhibitory domain to prevent KASH binding by modifying the homotrimeric conformation that is required for KASH binding or by directly blocking the KASH-binding site, while the region corresponding to the first coiled coil (CC1) may stabilize the trimer and act to suppress the inhibitory domain (Figure 6A).
Illustration of the SUN-KASH complex across the NE. (A) The N-terminal domain of SUN2 anchors into the INM, while the C-terminal SUN domain tethers the KASH domain to the ONM. The SUN domain trimer binds three separate KASH domains, forming a 3:3 hexameric heterocomplex. The two coiled-coil motifs CC1 and CC2 could function as activation and inhibitory domains, respectively, to regulate the (de)coupling of SUN-KASH assembly. (B) Under physiological conditions, the 3:3 SUN-KASH complex may undergo additional clustering, likely through hybrid (trimeric vs dimeric) intermolecular coiled-coils, to form higher-order networks in the luminal space of the NE, consistent with the mechano-transduction role of the LINC complex.
Our structural and biochemical studies strongly indicate that the SUN domain homotrimer is a primary KASH-binding unit, consistent with a recent independent report of the SUN-KASH complex structure33. However, it was recently found that both SUN proteins and KASH proteins can form high-order oligomers or clusters during specific cellular events23,34,35,36,37,38,39. Given that mechanical transduction is one of the salient features of LINC complex function, we have previously proposed and would like to reiterate that regulated networking could create a “machine” for the SUN-KASH-mediated force (de)coupling between the nucleus and the cytoskeleton (Figure 6B). While the second coiled-coil (CC2) and the SUN domain are trimeric, the first coiled-coil (CC1) of the SUN2 luminal domain is predicted to be dimeric12, suggesting potential hybrid networks between different SUN2 coiled-coil regions.
A study by Sosa et al. used sedimentation equilibrium analysis and suggested that the SUN complex was a homotrimer33. Nevertheless, large differences likely exist between the SUN-KASH microenvironment in cells and the solution conditions used for sedimentation equilibrium analysis. Under physiological conditions, the SUN-KASH assembly occurs in the luminal space of the nuclear double membranes, and the SUN and KASH proteins are anchored to the INM and ONM, respectively. These restrictions may facilitate and stabilize higher-order SUN-KASH networks. In addition, SUN proteins could form intermolecular disulfide bonds15 as well as bind to other yet to be identified proteins, raising the possibility of mechanical tuning across the NE through regulation of the SUN-KASH network. The exact regulatory mechanism of the LINC complex warrants additional investigation in the future.
Novel and important functions of the LINC complex have recently been discovered. For example, SUN2 has lately been identified as an interferon-stimulated gene that is likely involved in antiviral innate immune responses40. Another study suggested that the levels of SUN proteins are reduced during human cytomegalovirus (HCMV) infection, which dramatically alters the architecture of the nuclear periphery41. As a consequence, the nucleus becomes permeable to large molecules, presumably facilitating the assembly and egress of virus. Therefore, alteration of the LINC complex by viral infection may affect many aspects of cell functions, such as migration and cytoskeleton remodeling.
Our structural and biochemical analysis identified a hydrophobic pocket (the BI-pocket) on the surface of the SUN domain homotrimer as the docking site of the KASH domain PPPT motif. Sequence alignment indicates that these structural elements contain highly conserved residues, which may represent a conserved structural mechanism of the LINC complex assembly and functional regulation. Proper assembly of the SUN-KASH complex, which could be regulated by SUN2 coiled-coil motifs, appears to be required for normal cell migration. Our structural studies provide a framework for future investigation of the functions of the LINC complex.
Materials and Methods
Cloning, protein expression and purification
The SUN domain of WT human SUN2 (amino acids 522-717) was expressed and purified as previously reported12. Briefly, the plasmid of pET21b-SUN2-SUN domain with a C-terminal 6xHis tag was transformed into an E.coli BL21 (DE3) strain. Protein expression was induced at 16 °C, and the protein was purified at 4 °C using a nickel affinity column followed by gel filtration. All of the mutations were generated with a QuikChange Site-Directed Mutagenesis kit (Stratagene) and verified by sequencing. Similar procedures were used for the expression and purification of the mutant forms of SUN domain. KASH (human Nesprin 2G, Ser6856-Thr6885) and KASHΔ (human Nesprin 2G, Ser6856-Gly6881) fragments were cloned into the pGEX-4T-1 vector. GST-KASH and GST-KASHΔ fusion proteins were purified using a GST affinity column followed by gel filtration.
Crystallization, data collection and structure determination
To grow crystals of the SUN domain in complex with the KASH peptide, the purified SUN domain proteins (3 mg/ml) were mixed with synthesized KASH peptides (SFYPMLRYTNGPPPT) at a molar ratio of 1:10 (protein:peptide) for 1 h at 4 °C. Crystallization was performed using the sitting-drop vapor diffusion method. Crystals were grown at 4 °C in a drop with 1 μl of the protein solution and 1 μl of the reservoir solution (50 mM MgCl2, 100 mM HEPES, pH 7.5, 6% PEG5000MME) with 19.5 mM Methyl-6-O-(N-heptylcarbamoyl)-a-D-glucopyranoside (HECAMEG) added. Diffraction data were collected from a flash-cooled crystal at 100 K at beamline BL17U of the Shanghai Synchrotron Radiation Facility (SSRF), China. The diffraction data were processed with HKL200042. The structure of the SUN-KASH complex was solved by the molecular replacement method using the program AutoMR in Phenix and the structure of the SUN domain as a search model43. The structure was refined using Refmac5, and model building was performed in Coot44,45. The statistics of data collection and model refinement are summarized in Table 1. The structural coordinates of the SUN-KASH complex were deposited in Protein Data Bank under accession number 4FI9.
Kinetics assay
The GST-KASH fusion protein was biotinylated in a buffer of 20 mM HEPES and 300 mM HEPES, pH 7.0. The interaction between the SUN domain protein and the GST-KASH fusion protein was determined by biolayer interferometry using an Octet Red 96 instrument (ForteBio Inc.). Specifically, the biotinylated GST-KASH at ∼50 μg/ml was loaded onto streptavidin-coated biosensors and incubated with WT and various mutant forms of SUN domain proteins. All of the binding data were collected at 30 °C. The experiments comprised five steps: (1) baseline acquisition, (2) protein loading onto the sensor, (3) second baseline acquisition, (4) association of the SUN domain proteins for the measurement of kon, and (5) dissociation of the SUN domain proteins for the measurement of koff. Five concentrations ranging from 25 to 400 nM of various SUN domain proteins were used for detection. Equilibrium dissociation constants (Kd) were calculated by the ratio of Koff to Kon.
GST pull-down assay
GST-KASH protein coupled to a Glutathione-Sepharose resin was mixed with different SUN2 proteins at 4 °C for 1 h in 20 mM HEPES and 150 mM KCl, pH 7.0 and washed three times. The input and output samples were boiled in SDS-PAGE loading buffer and loaded on a 12% SDS-PAGE gel for separation followed by Coomassie blue staining.
The amount of SUN2 protein used for input was equal. In order to express and purify various SUN2 proteins in E. coil, we tried a series of expression tags. Specifically, WT, del AA' and S641E SUN2 were fused with His-tag at C-termini and other mutants were fused with His-tag at N-termini. Therefore the molecular weight of the input varies between WT and different mutant forms of SUN2 due to distinct ways of tag fusion.
Cell migration assay
Ovcar-3 (Ovarian cancer) cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum. The Ovcar-3 cells were transfected with pcDNA3.1, Myc-FAK, full-length Flag-SUN2 and its mutants for 24 h. Cell transwell chambers were obtained from Corning Costar (Corning NY). RPMI 1640 supplemented with 10% fetal bovine serum was added to the bottom chamber (600 μl). Next, 1 × 105 Ovcar-3 cells were added to the top chamber in serum free RPMI-1640 (500 μl). After 24 h, cells that migrated through the membrane and to the bottom surface of the inserts were fixed with 4% formaldehyde for 10 min and stained with a Three-Step Stain Set (Richard-Allan Scientific).
Accession codes
References
Crisp M, Liu Q, Roux K, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Bio 2006; 172:41–53.
Ostlund C, Folker ES, Choi JC, et al. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J Cell Sci 2009; 122:4099–4108.
Lombardi ML, Lammerding J . Keeping the LINC: the importance of nucleocytoskeletal coupling in intracellular force transmission and cellular function. Biochem Soc Trans 2011; 39:1729–1734.
Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP . Nuclear lamins: building blocks of nuclear architecture. Genes Dev 2002; 16:533–547.
Tzur YB, Margalit A, Melamed-Book N, Gruenbaum Y . Matefin/SUN-1 is a nuclear envelope receptor for CED-4 during Caenorhabditis elegans apoptosis. Proc Natl Acad Sci USA 2006; 103:13397–13402.
Gob E, Schmitt J, Benavente R, Alsheimer M . Mammalian sperm head formation involves different polarization of two novel LINC complexes. PloS One 2010; 5:e12072.
Hale CM, Shrestha AL, Khatau SB, et al. Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys J 2008; 95:5462–5475.
Worman HJ, Bonne G . “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res 2007; 313:2121–2133.
Mounkes L, Kozlov S, Burke B, Stewart CL . The laminopathies: nuclear structure meets disease. Curr Opin Genet Dev 2003; 13:223–230.
Wang Q, Du X, Cai Z, Greene MI . Characterization of the structures involved in localization of the SUN proteins to the nuclear envelope and the centrosome. DNA Cell Biol 2006; 25:554–562.
Hodzic DM, Yeater DB, Bengtsson L, Otto H, Stahl PD . Sun2 is a novel mammalian inner nuclear membrane protein. J Bio Chem 2004; 279:25805–25812.
Zhou Z, Du X, Cai Z, et al. Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope. J Bio Chem 2012; 287:5317–5326.
Liang Y, Chiu PH, Yip KY, Chan SY . Subcellular localization of SUN2 is regulated by Lamin A and Rab5. PloS One 2011; 6:e 20507.
Turgay Y, Ungricht R, Rothballer A, et al. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J 2010; 29:2262–2275.
Lu W, Gotzmann J, Sironi L, et al. Sun1 forms immobile macromolecular assemblies at the nuclear envelope. BBA-Mol Cell Res 2008; 1783:2415–2426.
Lei K, Zhang X, Ding X, et al. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc Natl Acad Sci USA 2009; 106:10207–10212.
Starr DA, Han M . A genetic approach to study the role of nuclear envelope components in nuclear positioning. Novartis Found Symp 2005; 264:208–219.
Luxton GG, Gomes ER, Folker ES, Worman HJ, Gundersen GG . TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus 2011; 2:173–181.
Luxton G, Gomes ER, Folker ES, Vintinner E, Gundersen GG . Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 2010; 329:956–959.
Taranum S, Vaylann E, Meinke P, et al. LINC complex alterations in DMD and EDMD/CMT fibroblasts. Eur J of Cell Biol 2012; 91:614–628.
Zhang Q, Bethmann C, Worth NF, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet 2007; 16:2816–2833.
Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D . Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res 2008; 314:1892–1905.
Ketema M, Wilhelmsen K, Kuikman I, et al. Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin. J Cell Sci 2007; 120:3384–3394.
Padmakumar VC, Libotte T, Lu W, et al. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci 2005; 118:3419–3430.
Rashmi RN, Eckes B, Glockner G, et al. The nuclear envelope protein Nesprin-2 has roles in cell proliferation and differentiation during wound healing. Nucleus 2011; 3:172–186.
Schaller MD . Cellular functions of FAK kinases: insight into molecular mechanisms and novel functions. J Cell Sci 2010; 123:1007–1013.
Sieg DJ, Hauck CR, Schlaepfer DD . Required role of focal adhesion kinase (FAK) for integrin-stimulated cell migration. J Cell Sci 1999; 112:2677–2691.
Yu J, Lei K, Zhou M, et al. KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum Mol Genet 2011; 20:1061–1073.
Starr DA, Hermann GJ, Malone CJ, et al. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development 2001; 128:5039–5050.
Lee KK, Starr D, Cohen M, et al. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol Biol Cell 2002; 13:892–901.
Oda Y, Fukuda H . Dynamics of Arabidopsis SUN proteins during mitosis and their involvement in nuclear shaping. Plant J 2011; 66:629–641.
Malone CJ, Fixsen WD, Horvitz HR, Han M . UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development 1999; 126:3171–3181.
Sosa BA, Rothballer A, Kutay U, Schwartz TU . LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 2012; 149:1035–1047.
Chikashige Y, Haraguchi T, Hiraoka Y . Another way to move chromosomes. Chromosoma 2007; 116:497–505.
Schmitt J, Benavente R, Hodzic D, et al. Transmembrane protein Sun2 is involved in tethering mammalian meiotic telomeres to the nuclear envelope. Proc Natl Acad Sci USA 2007; 104:7426–7431.
Mans BJ, Anantharaman V, Aravind L, Koonin EV . Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 2004; 3:1612–1637.
Mislow JM, Holaska JM, Kim MS, et al. Nesprin-1alpha self-associates and binds directly to emerin and lamin A in vitro. FEBS Lett 2002; 525:135–140.
Chikashige Y, Tsutsumi C, Yamane M, et al. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 2006; 125:59–69.
Penkner A, Tang L, Novatchkova M, et al. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev Cell 2007; 12:873–885.
Schoggins JW, Wilson SJ, Panis M, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 2011; 472:481–485.
Buchkovich NJ, Maguire TG, Alwine JC . Role of the endoplasmic reticulum chaperone BiP, SUN domain proteins, and dynein in altering nuclear morphology during human cytomegalovirus infection. J Virol 2010; 84:7005–7017.
Otwinowski Z, Minor W . Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 1997; 276:307–326.
Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010; 66:213–221.
Emsley P, Lohkamp B, Scott WG, Cowtan K . Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010; 66:486–501.
Murshudov GN, Skubak P, Lebedev AA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 2011; 67:355–367.
Acknowledgements
We thank the staff at BL17U of the Shanghai Synchrotron Radiation Facility for their help with the data collection. This work was supported by the 973 program of the Ministry of Science and Technology of China (2010CB529701, 2012CB910204), the National Natural Science Foundation of China (30970566, 10979005, 30971647, 31171414) and Shanghai Committee of Science and Technology (11JC14140000). ZZ is a scholar of the Hundred Talents Program of the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, W., Shi, Z., Jiao, S. et al. Structural insights into SUN-KASH complexes across the nuclear envelope. Cell Res 22, 1440–1452 (2012). https://doi.org/10.1038/cr.2012.126
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2012.126