Abstract
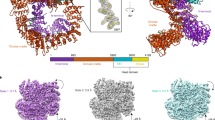
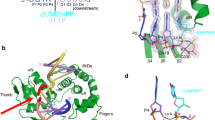
The Ku heterodimer (Ku70 and Ku80 subunits) contributes to genomic integrity through its ability to bind DNA double-strand breaks and facilitate repair by the non-homologous end-joining pathway. The crystal structure of the human Ku heterodimer was determined both alone and bound to a 55-nucleotide DNA element at 2.7 and 2.5 Å resolution, respectively. Ku70 and Ku80 share a common topology and form a dyad-symmetrical molecule with a preformed ring that encircles duplex DNA. The binding site can cradle two full turns of DNA while encircling only the central 3–4 base pairs (bp). Ku makes no contacts with DNA bases and few with the sugar-phosphate backbone, but it fits sterically to major and minor groove contours so as to position the DNA helix in a defined path through the protein ring. These features seem well designed to structurally support broken DNA ends and to bring the DNA helix into phase across the junction during end processing and ligation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
van Gent, D. C., Hoeijmakers, J. H. & Kanaar, R. Chromosomal stability and the DNA double-stranded break connection. Nature Rev. Genet. 2, 196–206 (2001).
Lieber, M. R. The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular eukaryotes. Genes Cells 4, 77–85 (1999).
Critchlow, S. E. & Jackson, S. P. DNA end-joining: from yeast to man. Trends Biochem. Sci. 23, 394–398 (1998).
Haber, J. E. Partners and pathways: repairing a double-strand break. Trends Genet. 16, 259–264 (2000).
Gottlieb, T. M. & Jackson, S. P. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72, 131–142 (1993).
Smith, G. C. & Jackson, S. P. The DNA-dependent protein kinase. Genes Dev. 13, 916–934 (1999).
Boulton, S. J. & Jackson, S. P. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15, 5093–5103 (1996).
Liang, F. & Jasin, M. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J. Biol. Chem. 271, 14405–14411 (1996).
Feldmann, E., Schmiemann, V., Goedecke, W., Reichenberger, S. & Pfeiffer, P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 28, 2585–2596 (2000).
Chen, S. et al. Accurate in vitro end-joining of a DNA double-strand break with partially cohesive 3′-overhangs and 3′-phosphoglycolate termini: effect of Ku on repair fidelity. J. Biol. Chem. 276, 24323–24330 (2001).
Taccioli, G. E. et al. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science 265, 1442–1445 (1994).
Zhu, C., Bogue, M. A., Lim, D. S., Hasty, P. & Roth, D. B. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell 86, 379–389 (1996).
Nussenzweig, A., Sokol, K., Burgman, P., Li, L. & Li, G. C. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival, and development. Proc. Natl Acad. Sci. USA 94, 13588–13593 (1997).
Gu, Y., Jin, S., Gao, Y., Weaver, D. T. & Alt, F. W. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc. Natl Acad. Sci. USA 94, 8076–8081 (1997).
Difilippantonio, M. J. et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 404, 510–514 (2000).
Ferguson, D. O. et al. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl Acad. Sci. USA 97, 6630–6633 (2000).
Roth, D. B. & Wilson, J. H. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol. Cell. Biol. 6, 4295–4304 (1986).
Thode, S., Schafer, A., Pfeiffer, P. & Vielmetter, W. A novel pathway of DNA end-to-end joining. Cell 60, 921–928 (1990).
Cary, R. B. et al. DNA looping by Ku and the DNA-dependent protein kinase. Proc. Natl Acad. Sci. USA 94, 4267–4272 (1997).
Ramsden, D. A. & Gellert, M. Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J. 17, 609–614 (1998).
Nick McElhinny, S. A., Snowden, C. M., McCarville, J. & Ramsden, D. A. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 20, 2996–3003 (2000).
Dynan, W. S. & Yoo, S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 26, 1551–1559 (1998).
Baumann, P. & West, S. C. DNA end-joining catalyzed by human cell-free extracts. Proc. Natl Acad. Sci. USA 95, 14066–14070 (1998).
Paillard, S. & Strauss, F. Site-specific proteolytic cleavage of Ku protein bound to DNA. Proteins 15, 330–337 (1993).
Singleton, B. K., Torres-Arzayus, M. I., Rottinghaus, S. T., Taccioli, G. E. & Jeggo, P. A. The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol. Cell. Biol. 19, 3267–3277 (1999).
Yoo, S., Kimzey, A. & Dynan, W. S. Photocross-linking of an oriented DNA repair complex. Ku bound at a single DNA end. J. Biol. Chem. 274, 20034–20039 (1999).
Kong, X.-P., Onrust, R., O'Donnell, M. & Kuriyan, J. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell 69, 425–437 (1992).
Gell, D. & Jackson, S. P. Mapping of protein-protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res. 27, 3494–4502 (1999).
Aravind, L. & Koonin, E. V. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25, 112–114 (2000).
Ghosh, G., van Duyne, G., Ghosh, S. & Sigler, P. B. Structure of NF-kappa B p50 homodimer bound to a kappa B site. Nature 373, 303–310 (1995).
Muller, C. W., Rey, F. A., Sodeoka, M., Verdine, G. L. & Harrison, S. C. Structure of the NF-kappa B p50 homodimer bound to DNA. Nature 373, 311–317 (1995).
de Vries, E., van Driel, W., Bergsma, W. G., Arnberg, A. C. & van der Vliet, P. C. HeLa nuclear protein recognizing DNA termini and translocating on DNA forming a regular DNA-multimeric protein complex. J. Mol. Biol. 208, 65–78 (1989).
Zhao, J., Wang, J., Chen, D. J., Peterson, S. R. & Trewhella, J. The solution structure of the DNA double-stranded break repair protein Ku and its complex with DNA: a neutron contrast variation study. Biochemistry 38, 2152–2159 (1999).
Giffin, W. et al. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature 380, 265–268 (1996).
Htun, H. & Dahlberg, J. E. Topology and formation of triple-stranded H-DNA. Science 243, 1571–1576 (1989).
Yoo, S. & Dynan, W. S. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 27, 4679–4686 (1999).
Calsou, P. et al. The DNA-dependent protein kinase catalytic activity regulates DNA end processing by means of Ku entry into DNA. J. Biol. Chem. 274, 7848–7856 (1999).
Stewart, J., Hingorani, M. M., Kelman, Z. & O'Donnell, M. Mechanism of β clamp opening by the δ subunit of E. coli DNA polymerase III holoenzyme. J. Biol. Chem. 276, 19182–19189 (2001).
Chou, C. H., Wang, J., Knuth, M. W. & Reeves, W. H. Role of a major autoepitope in forming the DNA binding site of the p70 (Ku) antigen. J. Exp. Med. 175, 1677–1684 (1992).
Otwinoski, W. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta. Crystallogr. D 55, 849–861 (1999).
Brünger, A. T. et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
CCP4. The CCP4 suite: programs for X-ray crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991).
Acknowledgements
We thank M. Jasin and K. Marians for critical reading of the manuscript; J. Gulbis for assistance with baculovirus production; L. Berman and M. Becker for use of synchrotron facilities at NSLS and C. Heaton at CHESS; and P. Jeffrey for help with synchrotron data collection. This work was supported by grants to J.G. from the NIH, HHMI and Pew Scholars Program in the Biomedical Sciences.
Author information
Authors and Affiliations
Corresponding author
Additional information
The atomic coordinates have been deposited in the Protein Data Bank under accession numbers 1JEQ and 1JEY.
Supplementary information
Rights and permissions
About this article
Cite this article
Walker, J., Corpina, R. & Goldberg, J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412, 607–614 (2001). https://doi.org/10.1038/35088000
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35088000
This article is cited by
-
Structural role for DNA Ligase IV in promoting the fidelity of non-homologous end joining
Nature Communications (2024)
-
Xrcc5/Ku80 is required for the repair of DNA damage in fully grown meiotically arrested mammalian oocytes
Cell Death & Disease (2023)
-
Human DNA-dependent protein kinase activation mechanism
Nature Structural & Molecular Biology (2023)
-
CRISPR-Cas12a induced DNA double-strand breaks are repaired by multiple pathways with different mutation profiles in Magnaporthe oryzae
Nature Communications (2022)
-
Fibroblast growth factor signalling influences homologous recombination-mediated DNA damage repair to promote drug resistance in ovarian cancer
British Journal of Cancer (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.