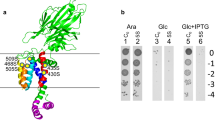
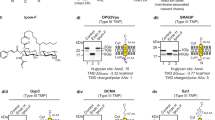
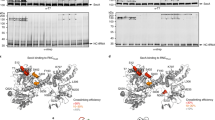
Abstract
The basic machinery for the translocation of proteins into or across membranes is remarkably conserved from Escherichia coli to humans. In eukaryotes, proteins are inserted into the endoplasmic reticulum using the signal recognition particle (SRP) and the SRP receptor, as well as the integral membrane Sec61 trimeric complex (composed of alpha, beta and gamma subunits)1. In bacteria, most proteins are inserted by a related pathway that includes the SRP homologue Ffh2,3,4,5, the SRP receptor FtsY6,7, and the SecYEG trimeric complex8, where Y and E are related to the Sec61 alpha and gamma subunits, respectively. Proteins in bacteria that exhibit no dependence on the Sec translocase were previously thought to insert into the membrane directly without the aid of a protein machinery9,10. Here we show that membrane insertion of two Sec-independent proteins requires YidC. YidC is essential for E. coli viability and homologues are present in mitochondria and chloroplasts. Depletion of YidC also interferes with insertion of Sec-dependent membrane proteins, but it has only a minor effect on the export of secretory proteins. These results provide evidence for an additional component of the translocation machinery that is specialized for the integration of membrane proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rapoport, T. A., Jungnickel, B. & Kutay, U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem. 65, 271–303 (1996).
Phillips, G. J. & Silhavy, T. J. The E. coli ffh gene is necessary for viability and efficient protein export. Nature 359, 744–746 ( 1992).
de Gier, J.-W. L. et al. Assembly of a cytoplasmic membrane protein in Escherichia coli is dependent on the signal recognition particle. FEBS Lett. 399, 307–309 ( 1996).
Bernstein, H. D., Zopf, D., Freyman, D. M. & Walter, P. Functional substitution of the signal recognition particle 54kD subunit by its Escherichia coli homolog. Proc. Natl Acad. Sci. USA 90, 5229–5233 (1993).
Ulbrandt, N. D., Newitt, J. A. & Bernstein, H. D. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88, 187–196 ( 1997).
Luirink, J. et al. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 13, 2289–2296 (1994).
Seluanov, A. & Bibi, E. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J. Biol. Chem. 272, 2053– 2055 (1997).
Wickner, W. & Leonard, M. R. Escherichia coli preprotein translocase. J. Biol. Chem. 22, 29514– 29516 (1996).
Geller, B. L. & Wickner, W. (1985). M13 procoat inserts into liposomes in the absence of other membrane proteins. J. Biol. Chem. 260, 13281– 13285.
de Gier, J.-W. L. et al. Differential use of the signal recognition particle translocase targeting pathway for inner membrane protein assembly in Escherichia coli . Proc. Natl Acad. Sci. USA 95, 14646 –14651 (1998).
Hell, K., Herrmann, J., Pratje, E., Neupert, W. & Stuart, R. A. Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett. 418, 367–370 (1997).
Hell, K., Herrmann, J. M., Pratje, E., Neupert, W. & Stuart, R. A. Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc. Natl Acad. Sci. USA 95, 2250–2255 (1998).
Sundberg, E. et al. ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell 9, 717–730 ( 1997).
Bonnefoy, N., Chalvet, F., Hamel, P., Slonimski, P. P. & Dujardin, G. Oxa1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J. Mol. Biol. 239, 201 –212 (1994).
Moore, M., Harrison, M. S., Peterson, E. C. & Henry, R. Chloroplast Oxa1p homolog Albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J. Biol. Chem. 275, 1529– 1532 (2000).
Stuart, R. A. & Neupert, W. Topogenesis of inner membrane proteins in mitochondria. Trends Biol. Sci. 21, 261 –267 (1996).
Glick, B. S. & von Heijne, G. Saccharomyces cerevisiae mitochondria lack a bacterial-type sec machinery. Protein Sci. 5, 2651–2652 ( 1996).
Whitley, P. et al. Sec-independent translocation of a 100-residue periplasmic N-terminal tail in the E. coli inner membrane protein proW. EMBO J. 13, 4653–4661 ( 1994).
Cristobal, S., Scotti, P., Luirink, J., von Heijne, G. & de Gier, J.-W. L. The signal recognition particle-targeting pathway does not necessarily deliver proteins to the Sec-translocase in Escherichia coli. J. Biol. Chem. 274, 20068– 20070 (1999).
Gilmore, R., Collins, P., Johnson, J., Kellaris, K. & Rapiejko, P. Transcription of full-length and truncated mRNA transcripts to study protein translocation across the endoplasmic reticulum. Methods Cell Biol. 34, 223–237 (1991).
Martoglio, B., Hofmann, M. W., Brunner, J. & Dobberstein, B. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell 18 , 207–214 (1995).
Scotti, P. A. et al. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19, 542–549 (2000).
Görlich, D., Hartmann, E., Prehn, S. & Rapoport, T. A. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature 357, 47–52 (1992).
Do, H., Falcone, D., Lin, J., Andrews, D. W. & Johnson, A. E. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell 85, 369–378 (1996).
Platt, R., Drescher, C. D., Park, S. K. & Phillips, G. J. Genetic system for reversible integration of DNA constructs and lacZ gene fusions into the Escherichia coli chromosome. Plasmid 43, 12–23 ( 2000).
Hamilton, C. M., Aldea, M., Washburn, B. K., Babitzke, P. & Kushner, S. R. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171, 4617–4622 ( 1989).
Graf, R., Brunner, J., Dobberstein, B. & Martoglio, B. in Cell Biology: a Laboratory Handbook 2nd edn, Vol. 4, (ed. Celis, J. E.) 495–501 (Academic, San Diego, 1998).
Brunner, J. Use of photocrosslinkers in cell biology. Trends Cell Biol. 6, 154–157 (1996).
Cload, S. T., Liu, D. R., Froland, W. A. & Schultz, P. Development of improved tRNAs for in vitro biosynthesis of proteins containing unnatural amino acids. Chem. Biol. 3, 1033–1038 (1996).
Gold, L. M. & Schweiger, M. in Methods in Enzymology Vol. 20, (eds Moldave, K. & Grossman, L.) 537– 542 (Academic, London and New York, 1971).
Acknowledgements
We thank P. G. Schultz for the E. coli suppressor tRNAAsn gene, S. R. Kushner for knockout vector pMAK705, J.-W. de Gier for the ProW construct, J. Brunner for the photocrosslinking reagent (Tmd)Phe-pdCpA, and Matthias Muller for advice with the amber suppression studies. This work was supported by an NSF grant (to R.E.D) and by a DFG grant (to A.K.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Samuelson, J., Chen, M., Jiang, F. et al. YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641 (2000). https://doi.org/10.1038/35020586
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35020586
This article is cited by
-
Cryo-EM structure of a bacteriophage M13 mini variant
Nature Communications (2023)
-
Dynamic feedback regulation for efficient membrane protein production using a small RNA-based genetic circuit in Escherichia coli
Microbial Cell Factories (2022)
-
Role of a bacterial glycolipid in Sec-independent membrane protein insertion
Scientific Reports (2022)
-
Structural and molecular mechanisms for membrane protein biogenesis by the Oxa1 superfamily
Nature Structural & Molecular Biology (2021)
-
Molecular communication of the membrane insertase YidC with translocase SecYEG affects client proteins
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.