Abstract
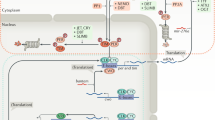
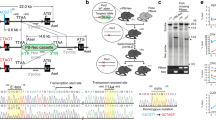
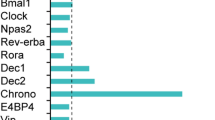
Circadian rhythms are driven by endogenous biological clocks that regulate many biochemical, physiological and behavioural processes in a wide range of life forms1. In mammals, there is a master circadian clock in the suprachiasmatic nucleus of the anterior hypothalamus. Three putative mammalian homologues (mPer1, mPer2 and mPer3) of the Drosophila circadian clock gene period (per) have been identified2,3,4,5,6,7,8. The mPer genes share a conserved PAS domain (a dimerization domain found in Per, Arnt and Sim) and show a circadian expression pattern in the suprachiasmatic nucleus. To assess the in vivo function of mPer2, we generated and characterized a deletion mutation in the PAS domain of the mouse mPer2 gene. Here we show that mice homozygous for this mutation display a shorter circadian period followed by a loss of circadian rhythmicity in constant darkness. The mutation also diminishes the oscillating expression of both mPer1 and mPer2 in the suprachiasmatic nucleus, indicating that mPer2 may regulate mPer1 in vivo. These data provide evidence that an mPer gene functions in the circadian clock, and define mPer2 as a component of the mammalian circadian oscillator.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pittendrigh, C. S. Temporal organization: reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 55, 16–54 (1993).
Sun, Z. S. et al. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell 90, 1003–1011 (1997).
Tei, H. et al. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature 389, 512–516 (1997).
Albrecht, U., Sun, Z. S., Eichele, G. & Lee, C. C. Adifferential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell 91, 1055–1064 (1997).
Shearman, L. P., Zylka, M. J., Weaver, D. R., Kolakowski, L. F. J & Reppert, S. M. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron 19, 1261–1269 (1997).
Takumi, T. et al. Anew mammalian period gene predominantly expressed in the suprachiasmatic nucleus. Genes Cells 3, 167–176 (1998).
Zylka, M. J., Shearman, L. P., Weaver, D. R. & Reppert, S. M. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20, 1103–1110 (1998).
Takumi, T. et al. Alight-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J. 17, 4753–4759 (1998).
Huang, Z. J., Edery, I. & Rosbash, M. PAS is a dimerization domain common to Drosophila Period and several transcription factors. Nature 364, 259–262 (1993).
Crosthwaite, S. K., Dunlap, J. C. & Loros, J. J. Neurospora wc-1 and wc-2 : transcription, photoresponses, and the origins of circadian rhythmicity. Science 276, 763–769 (1997).
King, D. P. et al. Positional cloning of the mouse circadian Clock gene. Cell 89, 641–653 (1997).
Rutila, J. E. et al. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93, 805–814 (1998).
Allada, R., White, N. E., So, W. V., Hall, J. C. & Rosbash, M. Amutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 93, 791–804 (1998).
Ponting, C. P. & Aravind, L. PAS: a multifunctional domain family comes to light. Curr. Biol. 7, R674–R677 (1997).
Bracewell, R. N. The Hartley Transform (Oxford Univ. Press, New York, (1986).
Aschoff, J. in Handbook of Behavioral Neurobiology 4: Biological Rhythms (ed. Aschoff, J.) 3–10 (Plenum, New York, (1981).
Ibuka, N., Inouye, S. I. & Kawamura, H. Analysis of sleep-wakefulness rhythms in male rats after suprachiasmatic nucleus lesions and ocular enucleation. Brain Res. 122, 33–47 (1977).
Vitaterna, M. H. et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264, 719–725 (1994).
Dowse, H. B. & Ringo, J. M. Further evidence that the circadian clock in Drosophila is a population of coupled ultradian oscillators. J. Biol. Rhythms 2, 65–76 (1987).
Hamblen-Coyle, M. J., Wheeler, D. A., Rutila, J. E., Rosbash, M. & Hall, J. C. Behavior of period-altered rhythm mutants of Drosophila in light:dark cycles. J. Insect Behav. 5, 417–446 (1992).
Ralph, M. R. & Menaker, M. Amutation of the circadian system in golden hamsters. Science 241, 1225–1227 (1988).
Hardin, P. E., Hall, J. C. & Rosbash, M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343, 536–540 (1990).
Dunlap, J. C. Genetics and molecular analysis of circadian rhythms. Annu. Rev. Genet. 30, 579–601 (1996).
Balsalobre, A., Damiola, F. & Schibler, U. Aserum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937 (1998).
Sangoram, A. M. et al. Mammalian circadian autoregulatory loop: A Timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron 21, 1101–1113 (1998).
Bae, K., Lee, C., Sidote, D., Chuang, K. Y. & Edery, I. Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM fucntion as positive regulators. Mol. Cell. Biol. 18, 6142–6151 (1998).
Matzuk, M. M., Finegold, M. J., Su, J. G., Hsueh, A. J. & Bradley, A. Alpha-inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature 360, 313–319 (1992).
Ramirez-Solis, R., Davis, A. C. & Bradley, A. Gene targeting in embryonic stem cells. Methods Enzymol. 225, 855–878 (1993).
Sokolove, P. G. & Bushell, W. N. The chi square periodogram: its utility for analysis of circadian rhythms. J. Theor. Biol. 72, 131–160 (1978).
Albrecht, U., Eichele, G., Helms, J. A. & Lu, H. in Molecular and Cellular Methods in Developmental Toxicology (ed. Daston, G. P.) 23–48 (CRC Press, Boca Raton, FL, (1997).
Acknowledgements
We thank S. Vaishnav, L. Qiu, Y.-C. Cheah, E. Sheppeard and S. Rivera for technical assistance; P. Hastings for comments on the manuscript; J. W. Patrick for providing space and facilities for circadian phenotype analysis; and J. Takahashi, Y. Zhao and M. Bucan for helpful discussions. This work was supported by grants from NINDS and NIDA to J. W. Patrick, from the Max-Planck Society to G. E., from NIH and the Department of Defense to C.C.L., and from NIH and the Howard Hughes Medical Institute to A.B. A.B. is an investigator with HHMI.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zheng, B., Larkin, D., Albrecht, U. et al. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 400, 169–173 (1999). https://doi.org/10.1038/22118
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/22118
This article is cited by
-
Knockout of the circadian gene, Per2, disrupts corticosterone secretion and results in depressive‐like behaviors and deficits in startle responses
BMC Neuroscience (2021)
-
Multiplexed CRISPR-Cas9 system in a single adeno-associated virus to simultaneously knock out redundant clock genes
Scientific Reports (2021)
-
Deletion of the clock gene Period2 (Per2) in glial cells alters mood-related behavior in mice
Scientific Reports (2021)
-
Modeling circadian regulation of ovulation timing: age-related disruption of estrous cyclicity
Scientific Reports (2020)
-
Time after time: circadian clock regulation of intestinal stem cells
Cellular and Molecular Life Sciences (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.