Abstract
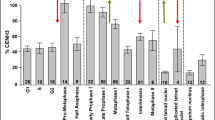
The organization of centromeric chromatin of diploid barley (Hordeum vulgare) encoding two (α and β) CENH3 variants was analysed by super-resolution microscopy. Antibody staining revealed that both CENH3 variants are organized in distinct but intermingled subdomains in interphase, mitotic and meiotic centromeres. Artificially extended chromatin fibres illustrate that these subdomains are formed by polynucleosome clusters. Thus, a CENH3 variant-specific loading followed by the arrangement into specific intermingling subdomains forming the centromere region appears. The CENH3 composition and transcription vary among different tissues. In young embryos, most interphase centromeres are composed of both CENH3 variants, while in meristematic root cells, a high number of nuclei contain βCENH3 mainly dispersed within the nucleoplasm. A similar distribution and no preferential arrangement of the two CENH3 variants in relationship to the spindle poles suggest that both homologs meet the same function in metaphase cells.




Similar content being viewed by others
Abbreviations
- CENH3:
-
Centromeric histone H3 variant
- CENP-C:
-
Centromere protein C
- SIM:
-
Structured illumination microscopy
References
Allshire RC, Karpen GH (2008) Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 9(12):923–937
Bai YW, Zhou Z, Feng HQ, Zhou BR (2011) Recognition of centromeric histone variant CenH3s by their chaperones. Structurally conserved or not. Cell Cycle 10(19):3217–3218. doi:10.4161/cc.10.19.17077
Blattner FR (2004) Phylogenetic analysis of Hordeum (Poaceae) as inferred by nuclear rDNA ITS sequences. Mol Phylogenet Evol 33(2):289–299
Blower MD, Sullivan BA, Karpen GH (2002) Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2(3):319–330
Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LE (2014) The quantitative architecture of centromeric chromatin. eLife 3:e02137. doi:10.7554/eLife.02137
Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL (2009) Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell 35(6):794–805. doi:10.1016/j.molcel.2009.07.022
Carroll CW, Milks KJ, Straight AF (2010) Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol 189(7):1143–1155. doi:10.1083/jcb.201001013
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162(1):156–159
Earnshaw WC, Allshire RC, Black BE, Bloom K, Brinkley BR, Brown W, Cheeseman IM, Choo KH, Copenhaver GP, Deluca JG, Desai A, Diekmann S, Erhardt S, Fitzgerald-Hayes M, Foltz D, Fukagawa T, Gassmann R, Gerlich DW, Glover DM, Gorbsky GJ, Harrison SC, Heun P, Hirota T, Jansen LE, Karpen G, Kops GJ, Lampson MA, Lens SM, Losada A, Luger K, Maiato H, Maddox PS, Margolis RL, Masumoto H, McAinsh AD, Mellone BG, Meraldi P, Musacchio A, Oegema K, O’Neill RJ, Salmon ED, Scott KC, Straight AF, Stukenberg PT, Sullivan BA, Sullivan KF, Sunkel CE, Swedlow JR, Walczak CE, Warburton PE, Westermann S, Willard HF, Wordeman L, Yanagida M, Yen TJ, Yoda K, Cleveland DW (2013) Esperanto for histones: CENP-A, not CenH3, is the centromeric histone H3 variant. Chromosome Res 21(2):101–106. doi:10.1007/s10577-013-9347-y
Heckmann S, Lermontova I, Berckmans B, De Veylder L, Baumlein H, Schubert I (2011) The E2F transcription factor family regulates CENH3 expression in Arabidopsis thaliana. Plant J 68(4):646–656. doi:10.1111/j.1365-313X.2011.04715.x
Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH (2006) Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell 10(3):303–315
Hirsch CD, Wu YF, Yan HH, Jiang JM (2009) Lineage-specific adaptive evolution of the centromeric protein CENH3 in diploid and allotetraploid Oryza species. Mol Biol Evol 26(12):2877–2885
Houben A, Schroeder-Reiter E, Nagaki K, Nasuda S, Wanner G, Murata M, Endo TR (2007) CENH3 interacts with the centromeric retrotransposon cereba and GC-rich satellites and locates to centromeric substructures in barley. Chromosoma 116(3):275–283
Jasencakova Z, Meister A, Schubert I (2001) Chromatin organization and its relation to replication and histone acetylation during the cell cycle in barley. Chromosoma 110(2):83–92
Kagawa N., Hori T, Yuko Hoki Y, Hosoya O, Tsutsui K, Saga Y, Sado T, Fukagawa T (2014) The CENPO complex requirement varis among different cell types. Chromosome Res 22 (3): 293–303
Kawabe A, Nasuda S, Charlesworth D (2006) Duplication of centromeric histone H3 (HTR12) gene in Arabidopsis halleri and A. lyrata, plant species with multiple centromeric satellite sequences. Genetics 174(4):2021–2032
Lermontova I, Schubert V, Fuchs J, Klatte S, Macas J, Schubert I (2006) Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell 18(10):2443–2451
Lermontova I, Kuhlmann M, Friedel S, Rutten T, Heckmann S, Sandmann M, Demidov D, Schubert V, Schubert I (2013) Arabidopsis KINETOCHORE NULL2 is an upstream component for centromeric histone H3 Variant cenH3 deposition at centromeres. Plant Cell 25(9):3389–3404. doi:10.1105/tpc.113.114736
Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature 389(6648):251–260
Marshall OJ, Chueh AC, Wong LH, Choo KH (2008) Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet 82(2):261–282. doi:10.1016/j.ajhg.2007.11.009
Moraes IC, Lermontova I, Schubert I (2011) Recognition of A. thaliana centromeres by heterologous CENH3 requires high similarity to the endogenous protein. Plant Mol Biol 75(3):253–261. doi:10.1007/s11103-010-9723-3
Nagaki K, Kashihara K, Murata M (2009) A centromeric DNA sequence colocalized with a centromere-specific histone H3 in tobacco. Chromosoma 118(2):249–257
Neumann P, Navratilova A, Schroeder-Reiter E, Koblizkova A, Steinbauerova V, Chocholova E, Novak P, Wanner G, Macas J (2012) Stretching the rules: monocentric chromosomes with multiple centromere domains. PLoS Genet 8(6):e1002777. doi:10.1371/journal.pgen.1002777
Ori A, Banterle N, Iskar M, Andres-Pons A, Escher C, Khanh Bui H, Sparks L, Solis-Mezarino V, Rinner O, Bork P, Lemke EA, Beck M (2013) Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol Syst Biol 9:648. doi:10.1038/msb.2013.4
Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci U S A 101(26):9903–9908. doi:10.1073/pnas.0307901101
Sanei M, Pickering R, Kumke K, Nasuda S, Houben A (2011) Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc Natl Acad Sci U S A 108(33):E498–E505. doi:10.1073/pnas.1103190108
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3(6):1101–1108
Schroeder-Reiter E, Sanei M, Houben A, Wanner G (2012) Current SEM techniques for de- and re-construction of centromeres to determine 3D CENH3 distribution in barley mitotic chromosomes. J Microsc 246(1):96–106. doi:10.1111/j.1365-2818.2011.03592.x
Shang WH, Hori T, Martins NMC, Toyoda A, Misu S, Monma N, Hiratani I, Maeshima K, Ikeo K, Fujiyama A, Kimura H, Earnshaw WC, Fukagawa T (2013) Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev Cell 24(6):635–648. doi:10.1016/j.devcel.2013.02.009
Shelden E, Wadsworth P (1993) Observation and quantification of individual microtubule behavior in vivo: microtubule dynamics are cell-type specific. J Cell Biol 120(4):935–945
Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11(11):1076–1083
Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR (2001) Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci 114(Pt 19):3529–3542
Acknowledgments
This work was supported by the German Federal Ministry of Education and Research (FKZ 0315965). We are grateful to Oda Weiß, Katrin Kumke and Karla Meier for excellent technical assistance and to Karin Lipfert for help with artwork.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Hans de Jong.
Takayoshi Ishii and Raheleh Karimi-Ashtiyani contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rotating barley metaphase chromosomes showing the centromeric localization of α and βCENH3. (AVI 4910 kb)
Enlargment of the centromeric region shown in Suppl. Movie 1. (AVI 5970 kb)
ESM 1
(DOC 117 kb)
Rights and permissions
About this article
Cite this article
Ishii, T., Karimi-Ashtiyani, R., Banaei-Moghaddam, A.M. et al. The differential loading of two barley CENH3 variants into distinct centromeric substructures is cell type- and development-specific. Chromosome Res 23, 277–284 (2015). https://doi.org/10.1007/s10577-015-9466-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-015-9466-8