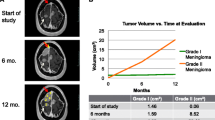
Abstract
Progressive meningiomas that have failed surgery and radiation have a poor prognosis and no standard therapy. While meningiomas are more common in females overall, progressive meningiomas are enriched in males. We performed a comprehensive molecular characterization of 169 meningiomas from 53 patients with progressive/high-grade tumors, including matched primary and recurrent samples. Exome sequencing in an initial cohort (n = 24) detected frequent alterations in genes residing on the X chromosome, with somatic intragenic deletions of the dystrophin-encoding and muscular dystrophy-associated DMD gene as the most common alteration (n = 5, 20.8%), along with alterations of other known X-linked cancer-related genes KDM6A (n =2, 8.3%), DDX3X, RBM10 and STAG2 (n = 1, 4.1% each). DMD inactivation (by genomic deletion or loss of protein expression) was ultimately detected in 17/53 progressive meningioma patients (32%). Importantly, patients with tumors harboring DMD inactivation had a shorter overall survival (OS) than their wild-type counterparts [5.1 years (95% CI 1.3–9.0) vs. median not reached (95% CI 2.9–not reached, p = 0.006)]. Given the known poor prognostic association of TERT alterations in these tumors, we also assessed for these events, and found seven patients with TERT promoter mutations and three with TERT rearrangements in this cohort (n = 10, 18.8%), including a recurrent novel RETREG1–TERT rearrangement that was present in two patients. In a multivariate model, DMD inactivation (p = 0.033, HR = 2.6, 95% CI 1.0–6.6) and TERT alterations (p = 0.005, HR = 3.8, 95% CI 1.5–9.9) were mutually independent in predicting unfavorable outcomes. Thus, DMD alterations identify a subset of progressive/high-grade meningiomas with worse outcomes.







Similar content being viewed by others
References
Arbajian E, Puls F, Antonescu CR, Amary F, Sciot R, Debiec-Rychter M et al (2017) In-depth genetic analysis of sclerosing epithelioid fibrosarcoma reveals recurrent genomic alterations and potential treatment targets. Clin Cancer Res 23:7426–7434. https://doi.org/10.1158/1078-0432.CCR-17-1856
Babadi Mehrtash DIB, Lee Samuel K, Andrey Smirnov AC, Lichtenstein Lee et al (2017) Abstract 3580: gATK CNV: copy-number variation discovery from coverage data. Am Assoc Cancer Res 77(13 Supplement):3580. https://doi.org/10.1158/1538-7445.AM2017-3580
Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J et al (2010) The landscape of somatic copy-number alteration across human cancers. Nature 463:899–905. https://doi.org/10.1038/nature08822
Bi WL, Greenwald NF, Abedalthagafi M, Wala J, Gibson WJ, Agarwalla PK et al (2017) Genomic landscape of high-grade meningiomas. NPJ Genom Med. https://doi.org/10.1038/s41525-017-0014-7
Blitshteyn S, Crook JE, Jaeckle KA (2008) Is there an association between meningioma and hormone replacement therapy? J Clin Oncol 26:279–282. https://doi.org/10.1200/JCO.2007.14.2133
Boström J, Meyer-Puttlitz B, Wolter M, Blaschke B, Weber RG, Lichter P et al (2001) Alterations of the tumor suppressor genes CDKN2A (p16(INK4a)), p14(ARF), CDKN2B (p15(INK4b)), and CDKN2C (p18(INK4c)) in atypical and anaplastic meningiomas. Am J Pathol 159:661–669. https://doi.org/10.1016/S0002-9440(10)61737-3
Brastianos PK, Horowitz PM, Santagata S, Jones RT, McKenna A, Getz G et al (2013) Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet 45:285–289. https://doi.org/10.1038/ng.2526
Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T et al (2012) Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 30:413–421. https://doi.org/10.1038/nbt.2203
Chamberlain JS, Metzger J, Reyes M, Townsend D, Faulkner JA (2007) Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. FASEB J 21:2195–2204. https://doi.org/10.1096/fj.06-7353com
Chamberlain MC (2012) The role of chemotherapy and targeted therapy in the treatment of intracranial meningioma. Curr Opin Oncol 24:666–671. https://doi.org/10.1097/CCO.0b013e328356364d
Cheng J, Demeulemeester J, Wedge DC, Vollan HKM, Pitt JJ, Russnes HG et al (2017) Pan-cancer analysis of homozygous deletions in primary tumours uncovers rare tumour suppressors. Nat Commun 8:1221. https://doi.org/10.1038/s41467-017-01355-0
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C et al (2013) Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31:213–219. https://doi.org/10.1038/nbt.2514
Cibulskis K, McKenna A, Fennell T, Banks E, DePristo M, Getz G (2011) ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics 27:2601–2602. https://doi.org/10.1093/bioinformatics/btr446
Clark VE, Erson-Omay EZ, Serin A, Yin J, Cotney J, Ozduman K et al (2013) Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science 339:1077–1080. https://doi.org/10.1126/science.1233009
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. https://doi.org/10.1093/bioinformatics/bts635
Ervasti JM (2007) Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim Biophys Acta 1772:108–117. https://doi.org/10.1016/j.bbadis.2006.05.010
Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM et al (2011) A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol 12:R1. https://doi.org/10.1186/gb-2011-12-1-r1
Flanigan KM, Dunn DM, von Niederhausern A, Soltanzadeh P, Gappmaier E, Howard MT et al (2009) Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum Mutat 30:1657–1666. https://doi.org/10.1002/humu.21114
Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W et al (2009) Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 27:182–189. https://doi.org/10.1038/nbt.1523
Goutagny S, Nault JC, Mallet M, Henin D, Rossi JZ, Kalamarides M (2014) High incidence of activating TERT promoter mutations in meningiomas undergoing malignant progression. Brain Pathol 24:184–189. https://doi.org/10.1111/bpa.12110
Goutagny S, Yang HW, Zucman-Rossi J, Chan J, Dreyfuss JM, Park PJ et al (2010) Genomic profiling reveals alternative genetic pathways of meningioma malignant progression dependent on the underlying NF2 status. Clin Cancer Res 16:4155–4164. https://doi.org/10.1158/1078-0432.CCR-10-0891
Harmancı AS, Youngblood MW, Clark VE, Coşkun S, Henegariu O, Duran D et al (2017) Integrated genomic analyses of de novo pathways underlying atypical meningiomas. Nat Commun 8:14433. https://doi.org/10.1038/ncomms14433
James MF, Stivison E, Beauchamp R, Han S, Li H, Wallace MR et al (2012) Regulation of mTOR complex 2 signaling in neurofibromatosis 2-deficient target cell types. Mol Cancer Res 10:649–659. https://doi.org/10.1158/1541-7786.MCR-11-0425-T
Juratli TA, Thiede C, Koerner MVA, Tummala SS, Daubner D, Shankar GM et al (2017) Intratumoral heterogeneity and TERT promoter mutations in progressive/higher-grade meningiomas. Oncotarget 8:109228–109237. https://doi.org/10.18632/oncotarget.22650
Jääskeläinen J, Haltia M, Servo A (1986) Atypical and anaplastic meningiomas: radiology, surgery, radiotherapy, and outcome. Surg Neurol 25:233–242
Kane AJ, Sughrue ME, Rutkowski MJ, Shangari G, Fang S, McDermott MW et al (2011) Anatomic location is a risk factor for atypical and malignant meningiomas. Cancer 117:1272–1278. https://doi.org/10.1002/cncr.25591
Katz LM, Hielscher T, Liechty B, Silverman J, Zagzag D, Sen R et al (2018) Loss of histone H3K27me3 identifies a subset of meningiomas with increased risk of recurrence. Acta Neuropathol 135:955–963. https://doi.org/10.1007/s00401-018-1844-9
Koelsche C, Sahm F, Capper D, Reuss D, Sturm D, Jones DT et al (2013) Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol 126:907–915. https://doi.org/10.1007/s00401-013-1195-5
Krayenbühl N, Pravdenkova S, Al-Mefty O (2007) De novo versus transformed atypical and anaplastic meningiomas: comparisons of clinical course, cytogenetics, cytokinetics, and outcome. Neurosurgery 61:495–503. https://doi.org/10.1227/01.neu.0000290895.92695.22 (discussion 503–494)
Kunkel LM, Hejtmancik JF, Caskey CT, Speer A, Monaco AP, Middlesworth W et al (1986) Analysis of deletions in DNA from patients with Becker and Duchenne muscular dystrophy. Nature 322:73–77. https://doi.org/10.1038/322073a0
Körner H, Epanchintsev A, Berking C, Schuler-Thurner B, Speicher MR, Menssen A et al (2007) Digital karyotyping reveals frequent inactivation of the dystrophin/DMD gene in malignant melanoma. Cell Cycle 6:189–198. https://doi.org/10.4161/cc.6.2.3733
Lalic T, Vossen RH, Coffa J, Schouten JP, Guc-Scekic M, Radivojevic D et al (2005) Deletion and duplication screening in the DMD gene using MLPA. Eur J Hum Genet 13:1231–1234. https://doi.org/10.1038/sj.ejhg.5201465
Lallemand D, Curto M, Saotome I, Giovannini M, McClatchey AI (2003) NF2 deficiency promotes tumorigenesis and metastasis by destabilizing adherens junctions. Genes Dev 17:1090–1100. https://doi.org/10.1101/gad.1054603
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. https://doi.org/10.1093/bioinformatics/btt656
Louis DN OH, Wiestler OD, Cavenee WK, Ellison DW, Figarella-Branger D, Perry A et al (2016) WHO classification of tumours of the central nervous system, vol 1. Revised 4th Edition
Luce LN, Abbate M, Cotignola J, Giliberto F (2017) Non-myogenic tumors display altered expression of dystrophin (DMD) and a high frequency of genetic alterations. Oncotarget 8:145–155. https://doi.org/10.18632/oncotarget.10426
McAvoy S, Ganapathiraju S, Perez DS, James CD, Smith DI (2007) DMD and IL1RAPL1: two large adjacent genes localized within a common fragile site (FRAXC) have reduced expression in cultured brain tumors. Cytogenet Genome Res 119:196–203. https://doi.org/10.1159/000112061
Perry A, Scheithauer BW, Stafford SL, Lohse CM, Wollan PC (1999) “Malignancy” in meningiomas: a clinicopathologic study of 116 patients, with grading implications. Cancer 85:2046–2056
Peyre M, Gauchotte G, Giry M, Froehlich S, Pallud J, Graillon T et al (2017) De novo and secondary anaplastic meningiomas: a study of clinical and histomolecular prognostic factors. Neuro Oncol. https://doi.org/10.1093/neuonc/nox231
Prins KW, Humston JL, Mehta A, Tate V, Ralston E, Ervasti JM (2009) Dystrophin is a microtubule-associated protein. J Cell Biol 186:363–369. https://doi.org/10.1083/jcb.200905048
Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G et al (2015) Oncotator: cancer variant annotation tool. Hum Mutat 36:E2423–E2429. https://doi.org/10.1002/humu.22771
Rempel SA, Schwechheimer K, Davis RL, Cavenee WK, Rosenblum ML (1993) Loss of heterozygosity for loci on chromosome 10 is associated with morphologically malignant meningioma progression. Cancer Res 53:2386–2392
Reuss DE, Piro RM, Jones DT, Simon M, Ketter R, Kool M et al (2013) Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol 125:351–358. https://doi.org/10.1007/s00401-013-1093-x
Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L et al (2012) Novel mutations target distinct subgroups of medulloblastoma. Nature 488:43–48. https://doi.org/10.1038/nature11213
Sahm F, Schrimpf D, Olar A, Koelsche C, Reuss D, Bissel J et al (2016) TERT promoter mutations and risk of recurrence in meningioma. J Natl Cancer Inst. https://doi.org/10.1093/jnci/djv377
Sahm F, Schrimpf D, Stichel D, Jones DTW, Hielscher T, Schefzyk S et al (2017) DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol 18:682–694. https://doi.org/10.1016/S1470-2045(17)30155-9
Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK (2012) Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28:1811–1817. https://doi.org/10.1093/bioinformatics/bts271
Shankar GM, Abedalthagafi M, Vaubel RA, Merrill PH, Nayyar N, Gill CM et al (2017) Germline and somatic BAP1 mutations in high-grade rhabdoid meningiomas. Neuro Oncol 19:535–545. https://doi.org/10.1093/neuonc/now235
Shaw RJ, McClatchey AI, Jacks T (1998) Localization and functional domains of the neurofibromatosis type II tumor suppressor, merlin. Cell Growth Differ 9:287–296
Shaw RJ, McClatchey AI, Jacks T (1998) Regulation of the neurofibromatosis type 2 tumor suppressor protein, merlin, by adhesion and growth arrest stimuli. J Biol Chem 273:7757–7764
Smith KM, Fagan PC, Pomari E, Germano G, Frasson C, Walsh C et al (2018) Antitumor activity of entrectinib, a Pan-TRK, ROS1, and ALK inhibitor, in. Mol Cancer Ther 17:455–463. https://doi.org/10.1158/1535-7163.MCT-17-0419
Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT et al (2011) Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 333:1039–1043. https://doi.org/10.1126/science.1203619
Strickland MR, Gill CM, Nayyar N, D’Andrea MR, Thiede C, Juratli TA et al (2016) Targeted sequencing of SMO and AKT1 in anterior skull base meningiomas. J Neurosurg. https://doi.org/10.3171/2016.8.jns161076
Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P (2015) Sambamba: fast processing of NGS alignment formats. Bioinformatics 31:2032–2034. https://doi.org/10.1093/bioinformatics/btv098
Taylor LE, Kaminoh YJ, Rodesch CK, Flanigan KM (2012) Quantification of dystrophin immunofluorescence in dystrophinopathy muscle specimens. Neuropathol Appl Neurobiol 38:591–601. https://doi.org/10.1111/j.1365-2990.2012.01250.x
Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F et al (2015) The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood 125:13–21. https://doi.org/10.1182/blood-2014-05-577270
Wang Y, Fletcher JA (2015) Cell cycle and dystrophin dysregulation in GIST. Cell Cycle 14:2713–2714. https://doi.org/10.1080/15384101.2015.1064676
Wang Y, Marino-Enriquez A, Bennett RR, Zhu M, Shen Y, Eilers G et al (2014) Dystrophin is a tumor suppressor in human cancers with myogenic programs. Nat Genet 46:601–606. https://doi.org/10.1038/ng.2974
Weber RG, Boström J, Wolter M, Baudis M, Collins VP, Reifenberger G et al (1997) Analysis of genomic alterations in benign, atypical, and anaplastic meningiomas: toward a genetic model of meningioma progression. Proc Natl Acad Sci USA 94:14719–14724
White SJ, den Dunnen JT (2006) Copy number variation in the genome; the human DMD gene as an example. Cytogenet Genome Res 115:240–246. https://doi.org/10.1159/000095920
Wigertz A, Lönn S, Mathiesen T, Ahlbom A, Hall P, Feychting M et al (2006) Risk of brain tumors associated with exposure to exogenous female sex hormones. Am J Epidemiol 164:629–636. https://doi.org/10.1093/aje/kwj254
Zankl H, Seidel H, Zang KD (1975) Cytological and cytogenetical studies on brain tumors. V. Preferential loss of sex chromosomes in human meningiomas. Humangenetik 27:119–128
Zhang NR, Siegmund DO, Ji H, Li JZ (2010) Detecting simultaneous changepoints in multiple sequences. Biometrika 97:631–645. https://doi.org/10.1093/biomet/asq025
Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD et al (2014) Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 20:1479–1484. https://doi.org/10.1038/nm.3729
Acknowledgements
This work is supported by US National Institutes of Health (NIH) 1R21NS099844 (to Drs. D. Cahill and P. Brastianos); the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Projektnummer: 401837860 and the Max Kade Foundation (to Dr. T. Juratli); the Meningioma Mommas (to Dr. H. Wakimoto); the Burroughs Wellcome Fund Career Award (to Dr. D. Cahill.); the Brain Science Foundation, the American Brain Tumor Association, and the Damon Runyon Research Foundation (to Dr. P. Brastianos).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
I.M.S. and H.A.E. are employees in Ignyta, and Inc. J.C. is an employee and owns stocks in Ignyta, Inc. All other authors have no conflicts of interest to report with regard to this manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1:
Detailed method and materials description: Whole-exome sequencing, copy number analysis, RNA sequencing, Multiplex Ligation-dependent Probe Amplification (MLPA), Western blotting, Electron Microscopy and Immunofluorescence (PDF 540 kb)
Online Resource 2: Supplementary Table
1: patients’ and tumor characteristics (XLSX 19 kb)
Online Resource 3:
Supplementary Fig. 1 | Copy number variations in progressive/high-grade meningiomas Deletions are depicted in blue, whereas amplifications are shown in red. Supplementary Fig. 2 | An overview of DMD deletions across multiple GISTIC analyses performed on the entire dataset (all_cancers) DMD is significantly focally deleted in 13 of 33 independent cancer types analyzed in the dataset. Among these, it is located within a focal peak region of deletion in 10 cancer types. For reference, 11.221% of all genes are significantly focally deleted in at least 13 cancer types and 5.877% of all genes are present in focal deletion peaks in at least 10 cancer types. The q-value calculated of focal DMD deletions in our cohort is 10−9.(http://portals.broadinstitute.org/tcga/gistic/browseGisticByGene). Supplementary Fig. 3 | Myogenic transcriptomic signature in higher-grade/progressive meningiomas RNA expression data was analyzed using the GTEx portal (https://www.gtexportal.org/home/) to identify genes that are predominantly expressed in the mesodermal tissue (skeletal or heart muscles). The expression of these genes was compared between samples with and without DMD inactivation. Of the seven mesodermal genes with detectable expression in at least three samples, a markedly higher expression level of these genes was observed in meningioma samples with DMD deletions.The seven genes shown in the heat map are:- ATPase Sarcoplasmic/Endoplasmic Reticulum Ca2 + Transporting 1 (ATP2A1): encodes one of the SERCA Ca(2 +)-ATPases, which are intracellular pumps located in the sarcoplasmic or endoplasmic reticula of muscle cells.- Desmin (DES): encodes a muscle-specific class III intermediate filament.- Enolase 3 (ENO3): encodes an isoenzyme in skeletal muscle cells that may play a role in muscle development and regeneration.- Myosin light chain, phosphorylatable, fast skeletal muscle (MYLPF).- Titin-cap(TCAP): encodes a protein found in striated and cardiac muscle that binds to the titin Z1–Z2 domains and is a substrate of titin kinase, interactions thought to be critical to sarcomere assembly. Mutations in this gene are associated with limb-girdle muscular dystrophy type 2G- Tropomyosin (TPM2): encodes beta-tropomyosin, a member of the actin filament-binding protein family, and mainly expressed in slow, type 1 muscle fibers.- Troponin T1 (TNNT1): encodes a protein that is a subunit of troponin, which is a regulatory complex located on the thin filament of the sarcomere. Supplementary Fig. 4 | DMD inactivation in the malignant IOMM-Lee meningioma cell line(a) Immunohistochemistry with DYS1 demonstrates loss of dystrophin expression in IOMM-Lee cells implanted into the mouse brain. (b) Immunofluorescence using DMD ab15227 (Abcam) revealed the cytoplasmic loss of expression in the IOMM-Lee cell line when compared with the Ben-Men-1 (BM1) cell line. (c) Screenshot of DMD mRNA expression in three anaplastic meningioma cell lines (IOMM-Lee, CH157 and F5) as shown by the CCLE dataset. A significant reduction of DMD mRNA expression is seen in the IOMM-Lee cell line (RPKM 0.000712), whereas CH157 and F5 cell lines showed a higher expression (RPKM 0.404 and 0.510, respectively). https://portals.broadinstitute.org/ccle/data Supplementary Fig. 5 | Additional Western blotting, RT-PCR and electron microscopic data(a) Representative Western blotting using DYS1 (reacts with the rod domain of dystrophin) shows loss of dystrophin 427 kDa expression in two patient samples with DMD deletion (MGH001 and MGH025) and in the malignant IOMM-Lee meningioma cell line. (b) An example for a reverse transcriptase PCR demonstrating loss of DMD mRNA expression of exons 10–15 and 63–68 in case MGH031 (DMD-deleted sample), compared to samples of patients with intact DMD. (c) Representative electron microscopic imaging demonstrates the DMD-inactivated IOMM-Lee cell line displayed predominantly rounded cell morphology with an apparently lower density of cytoskeleton filaments compared with the human arachnoid cells (AC007-hTERT). (PDF 6619 kb)
Online Resource 4: Supplementary Table
2: Sheet 1: List of all synonymous and non-synonymous mutations identified using the criteria for somatic mutation calling Sheet 2: List of genes residing in late-replicating common fragile sites that were assessed for deletions in the current study Sheet 3: List of genes larger than 1 Mb evaluated for alterations in the current study (XLSX 498 kb)
Online Resource 5: Supplementary Table
3: Sheet 1: TERT fusion data Sheet 2: List of used RT-PCR primers in the study Sheet 3: List of used RT-PCR primers to validate the TERT fusions (XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Juratli, T.A., McCabe, D., Nayyar, N. et al. DMD genomic deletions characterize a subset of progressive/higher-grade meningiomas with poor outcome. Acta Neuropathol 136, 779–792 (2018). https://doi.org/10.1007/s00401-018-1899-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-018-1899-7