-
PDF
- Split View
-
Views
-
Cite
Cite
Matthew R. Northam, Elizabeth A. Moore, Tony M. Mertz, Sara K. Binz, Carrie M. Stith, Elena I. Stepchenkova, Kathern L. Wendt, Peter M. J. Burgers, Polina V. Shcherbakova, DNA polymerases ζ and Rev1 mediate error-prone bypass of non-B DNA structures, Nucleic Acids Research, Volume 42, Issue 1, 1 January 2014, Pages 290–306, https://doi.org/10.1093/nar/gkt830
Close - Share Icon Share
Abstract
DNA polymerase ζ (Pol ζ) and Rev1 are key players in translesion DNA synthesis. The error-prone Pol ζ can also participate in replication of undamaged DNA when the normal replisome is impaired. Here we define the nature of the replication disturbances that trigger the recruitment of error-prone polymerases in the absence of DNA damage and describe the specific roles of Rev1 and Pol ζ in handling these disturbances. We show that Pol ζ/Rev1-dependent mutations occur at sites of replication stalling at short repeated sequences capable of forming hairpin structures. The Rev1 deoxycytidyl transferase can take over the stalled replicative polymerase and incorporate an additional ‘C’ at the hairpin base. Full hairpin bypass often involves template-switching DNA synthesis, subsequent realignment generating multiply mismatched primer termini and extension of these termini by Pol ζ. The postreplicative pathway dependent on polyubiquitylation of proliferating cell nuclear antigen provides a backup mechanism for accurate bypass of these sequences that is primarily used when the Pol ζ/Rev1-dependent pathway is inactive. The results emphasize the pivotal role of noncanonical DNA structures in mutagenesis and reveal the long-sought-after mechanism of complex mutations that represent a unique signature of Pol ζ.
INTRODUCTION
DNA polymerases α, δ and ε perform the bulk of DNA synthesis during eukaryotic DNA replication (1,2). DNA damage resulting from exogenous and endogenous factors creates obstacles for the replicative enzymes. The bypass of replication impediments is facilitated by specialized translesion synthesis (TLS) polymerases (3). These include DNA polymerase ζ (Pol ζ), Pol η, Pol ι, Pol κ and Rev1 in humans. The yeast Saccharomyces cerevisiae has homologs of Pol ζ, Pol η and Rev1. TLS is a mutagenic process because of the miscoding potential of the damaged nucleotides and the inherently lower selectivity of the active sites of the specialized polymerases (4). In yeast and human cells, DNA synthesis by Pol ζ is responsible for nearly all mutagenesis induced by exogenous genotoxicants (5). Pol ζ is also required for the vast majority of mutations provoked by endogenous DNA damage [see references in (6)]. The specific role of Pol ζ in TLS is thought to be the extension of aberrant primer termini resulting from nucleotide incorporation opposite lesions by other DNA polymerases. Pol ζ is uniquely capable of extending poorly matched primer termini, including those containing nucleotides across from noncoding and helix-distorting lesions (3). During copying of undamaged DNA in vitro, Pol ζ also frequently misincorporates nucleotides showing overall low fidelity (7).
The deoxycytidyl transferase Rev1 is an essential partner of Pol ζ in TLS. Like Pol ζ, Rev1 is required for DNA damage-induced mutagenesis. Although the catalytic activity of Rev1 is used in vivo during the bypass of several lesions (8–12), the essential role of Rev1 in TLS is structural. Yeast Rev1 and its mammalian homologs are involved in multiple physical interactions with other TLS polymerases (5). In addition, Rev1 has ubiquitin-binding motifs through which it can interact with ubiquitylated proliferating cell nuclear antigen (PCNA) at stalled replication forks (13,14). This led to the idea that Rev1 could provide a docking site to help exchange different DNA polymerases at the replication fork. Binding of Rev1 to Pol ζ also stimulates the mismatch extension and TLS activity of Pol ζ (15).
In addition to the role of Pol ζ and Rev1 in TLS, multiple reports document Pol ζ-dependent mutagenesis in situations when cells are not expected to accumulate excessive DNA damage. The Pol ζ/Rev1-dependent processes are responsible for 50–70% of spontaneous mutations in wild-type S. cerevisiae strains (16). Pol ζ also contributes to an increased mutation rate associated with high levels of transcription (17) or double-strand break repair (18). We and others have shown that Pol ζ participation in replication and mutagenesis is promoted by a variety of replication machinery defects (6,19–21). For example, defects in S. cerevisiae replicative polymerases Pol δ and Pol ε that are thought to affect replication fork progression or the replisome integrity lead to a mutator phenotype. A total of 80–90% of spontaneous mutations in these strains are mediated by Pol ζ (6,20,21), a phenomenon that we termed defective-replisome-induced-mutagenesis (DRIM). Similar Pol ζ-dependent mutagenesis is observed after treatment of wild-type cells with the replication inhibitor hydroxyurea (22). Our recent studies provided evidence that DRIM results from the participation of Pol ζ in the copying of undamaged DNA rather than from mutagenic lesion bypass (22). The recruitment of Pol ζ for the replication of undamaged DNA was unexpected because the access of error-prone polymerases to the replication fork must be tightly regulated to restrict mutagenesis.
The exact nature of replication problems that signal for the recruitment of Pol ζ to undamaged templates remained enigmatic. This study provides evidence for a model wherein replication stalling at transient noncanonical DNA structures is a major factor activating the Pol ζ/Rev1-dependent mutagenic replication pathway. DNA sequences capable of adopting unusual secondary structures, such as hairpins, cruciforms, slipped structures, G-quadruplex, triplex and Z-DNA, are widespread in the eukaryotic genomes. Inverted repeat sequences, as well as certain tri- and tetranucleotide repeats, are prone to hairpin structure formation, particularly when the repeated sequence is exposed as a single-stranded DNA during DNA replication, repair or transcription (23–25). These sequences inhibit replication progression in bacteria, yeast and mammalian cells (26–30), as well as block DNA polymerases in the in vitro replication assays (31–35). These sequences also promote genomic instability (DNA breaks, mutations and gross rearrangements) (36–49). Genomic rearrangements triggered by hairpin and cruciform structures are thought to contribute to the pathogenesis of many human diseases (50,51). Importantly, the replication-blocking structures known to increase the genome instability are typically formed by repetitive sequences at least 50–100 and sometimes >1000-nt long. In contrast, hundreds of short (4–5 nt) repeats are present in the coding sequences of every gene and are not generally thought to present an obstacle for the DNA replication machinery. The role of the local DNA structure in mutagenesis in these regions, generally referred to as non-repetitive, is poorly understood. Genetic studies in bacteriophage T4 and Escherichia coli suggested that abnormal replication of short quasipalindromic sequences can lead to a high frequency of complex mutations leading to the perfection of the palindrome (46,47,52–56). Evidence for the abnormal replication progression through these sequences, however, is lacking. It is also not known whether such short inverted repeat sequences can form non-B DNA structures and cause genome instability in eukaryotic cells. Eukaryotic Pol ζ has long been known to generate complex mutations (multiple changes within short DNA stretches) at a high rate (7,22,57), but the mechanism of this unique type of mutation remained unclear.
In this study, we demonstrate that an unexpectedly high proportion of mutagenesis during replication is due to polymerase stalling at short DNA repeats capable of forming transient noncanonical DNA structures. We further implicate Pol ζ and Rev1 in the error-prone bypass of these structures via a template-switching type of DNA synthesis at the replication fork. We also describe elementary biochemical steps leading to complex mutation generation by Pol ζ/Rev1.
MATERIALS AND METHODS
Saccharomyces cerevisiae strains and plasmids
All S. cerevisiae strains used in this study are isogenic to the E134 strain [MATα ade5-1 lys::InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52; (58)]. The rev1Δ and mmsΔ derivatives of E134 were obtained by transformation with polymerase chain reaction-generated DNA fragments carrying a selectable kanMX cassette flanked by short sequence homology to REV1 or MMS2. The disruptions were confirmed by polymerase chain reaction and by ultraviolet-immutability (rev1Δ) or the mutator phenotype (mmsΔ) of the transformants. To replace the chromosomal REV1 gene with the rev1-1 and rev1-cd alleles, a URA3-based integrative plasmid pRS306-REV1 was constructed by ligating a DNA fragment containing the first 1818 nt of the REV1 open reading frame and 75 nt of the upstream sequence into SpeI and HindIII sites of pRS306 (59). The rev1-1 [Gly193→Arg; (60)] and rev1-cd (Asp467→Ala, Glu468→Ala) mutations were created in the pRS306-REV1 plasmid by site-directed mutagenesis using a QuickChange site-directed mutagenesis kit from Stratagene. Strain E134 was transformed by EcoRI-digested pRS306-rev1-1 and pRS306-rev1-cd, and Ura+ transformants were plated on a medium containing 5-fluoroorotic acid to select for excision of the plasmid. The REV1 allele replacement was confirmed by DNA sequencing. The pol3-Y708A mutants were constructed as described previously (20). During the construction of double rev1-cd pol3-Y708A and triple mmsΔ rev1-cd pol3-Y708A strains, the pol3-Y708A allele was always introduced last to limit the accumulation of additional random mutations in these genetically unstable strains.
The M13/lacZ::CAN1 (1-1560-F) and M13/lacZ::CAN1 (1-1560-R) plasmids were constructed by ligating a fragment of DNA containing the first 1560 bases of the yeast CAN1 gene in the forward and reverse orientations, respectively, into the EcoRI site in the lacZ gene of the M13mp2.
Measurement of the spontaneous mutation rate and mutational spectra analysis
The rate of spontaneous mutation to canavanine resistance (Canr) was measured by fluctuation analysis, and the mutational spectra were determined as previously described (22). The significance of differences between Canr mutation rates was estimated by using the Wilcoxon-Mann-Whitney nonparametric criterion (61).
Enzymes
The T4 Pol was purchased from New England Biolabs (NEB). The yeast Pol δ, Pol ζ, Rev1 and its catalytically inactive variant (Rev1-CD), PCNA, replicaton factor C (RFC) and replication protein A (RPA) were purified as described in (14,62–66) respectively.
In vitro DNA synthesis assays
Singly primed circular DNA substrates for DNA polymerase assays were prepared by annealing the M13/CAN1 (1-1560-F) or M13/lacZ::CAN1 (1-1560-R) ssDNA to Cy-5-labeled oligonucleotides complementary to different CAN1 regions. The M13/lacZ::CAN1 (1-1560-F) and M13/lacZ::CAN1 (1-1560-R) ssDNA was purified from E. coli cultures infected with the corresponding bacteriophages as described in (67). The standard purification procedure was followed by treatment with T4 Pol (NEB) and nuclease-free RNase A (USB) for 1 h at 37°C to remove any remaining regions of double-stranded DNA and RNA contaminants. High pressure liquid chromatography-purified Cy5-labeled oligonucleotide primers were purchased from IDT. The oligonucleotides were further purified by extraction from 18% polyacrylamide gel (68) and concentrated using Amicon ultra centrifugation columns (Millipore). Oligonucleotide primers used to study DNA polymerase pausing during copying of the CAN1 template are described in Supplementary Figure S2. Oligonucleotides used to generate substrates with multiple mismatched bases and fully matched control substrates are described in Figure 7. The annealing was achieved by incubating 20 µg (∼8 pmol) of M13/CAN1 (1-1560-F) or M13/lacZ::CAN1 (1-1560-R) with 2.4 pmol of Cy5-labeled oligonucleotide in the presence of 150 mM NaAc at 78°C for 2 min and then cooling slowly to room temperature (∼2 h). The reactions were desalted by spinning through Amicon ultra centrifugation columns (Millipore). The annealing was verified by electrophoresis in 0.8% Tris-acetate-EDTA (TAE) agarose gels, and the substrates were stored at −80°C.
DNA synthesis reactions with T4 Pol were performed at 37°C in NEB buffer 2 and contained 20 nM DNA substrate, 100 µM of each dNTP and either 2 or 0.2 nM T4 Pol as indicated. The reactions with yeast Pol δ, Pol ζ, Rev1 and Rev1-CD were performed at 30°C and contained 40 mM Tris–HCl (pH 7.8), 100 mM NaCl, 8 mM MgAc, 1 mM DTT, 200 µg/ml bovine serum albumine, 2.5% glycerol, 20 nM DNA substrate, 100 µM of each dNTP, 4.9 µM RPA, 0.006 µM RFC, 0.016 µM PCNA, 0.4 mM ATP and the indicated polymerase(s) at the following concentrations: Pol δ—0.4 nM, Rev1 and Rev1-CD—40 nM, Pol ζ—5 nM. Incubation was for 10 min unless stated otherwise. Reactions were stopped by placing the tubes on ice and adding 1.5 µl of 0.5 M EDTA. The reactions were then incubated with Proteinase K at 37°C for 10 min and purified by phenol/chloroform extraction. DNA was precipitated with ethanol and resuspended in formamide loading buffer. The reaction products were separated by electrophoresis in 12% denaturing polyacrylamide gel and detected and quantified using the Typhoon imaging system and ImageQuant software (GE Healthcare). The sequencing ladder markers for the polymerase pausing analysis were produced by using the same DNA templates and primers and Sequenase DNA Sequencing Kit (Affymetrix) according to the manufacturer’s instructions.
DNA secondary structure prediction
The CAN1 gene sequence was analyzed for the presence of potential secondary structures using the mfold Web Server provided by the RNA Institute at the College of Arts and Sciences, University at Albany, State University of New York (http://mfold.rna.albany.edu/?q=mfold/dna-folding-form) (69).
RESULTS
Both the structural and the catalytic functions of Rev1 contribute to DRIM
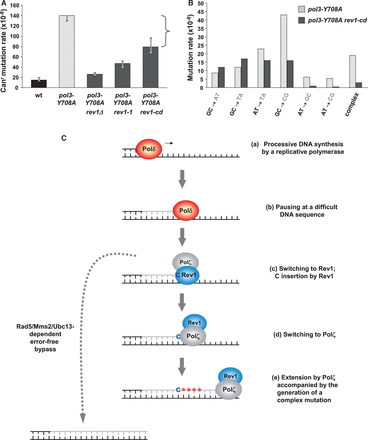
Many mutations affecting the yeast replicative DNA polymerases cause a Pol ζ-dependent spontaneous mutator phenotype that we refer to as DRIM. We previously demonstrated that the mechanism of DRIM involves impaired replication fork progression in the polymerase mutants (70) that leads to a constitutive monoubiquitination of PCNA (6) and the recruitment of Pol ζ for replication of undamaged DNA (22). To model DRIM in the present study, we use the pol3-Y708A mutation resulting in a single amino acid change in the active site of Pol δ. This mutation causes a moderately impaired growth, sensitivity to the replication inhibitor hydroxyurea, a robust monoubiquitination of PCNA and an increase in spontaneous mutagenesis that is almost entirely Pol ζ-dependent (6,20). The spectrum of spontaneous mutations in the pol3-Y708A strain shows the predominance of base substitutions with a particularly high proportion of GC→CG transversions, a low frequency of frameshifts and a high frequency of complex mutations (22). The complex mutations are defined as multiple changes within short (up to 6 nt) DNA stretches. We refer to them as type I complex mutations to distinguish them from more dramatic sequence substitutions that are seen at a low rate in Pol ζ-deficient strains [(22) and discussion below]. This spectrum closely resembles the error specificity of Pol ζ during copying of undamaged DNA in vitro (7), suggesting that the mutator effect of pol3-Y708A results from the error-prone DNA synthesis by Pol ζ.
The present study initially aimed to characterize the role of Rev1 in DRIM. Because of the essential role of the Rev1 protein in many Pol ζ-dependent transactions, we hypothesized that Rev1 performs a structural function in DRIM as well. Indeed, a deletion of the REV1 gene reduced the rate of spontaneous mutation to canavanine resistance (Canr) in the pol3-Y708A strain to nearly a wild-type level (Figure 1A and Supplementary Table S1), similar to the effect of REV3 deletion (6). Comparison of the spectra of can1 mutations in the pol3-Y708A rev1Δ and pol3-Y708A rev3Δ strains showed that the two spectra were indistinguishable, and the physical presence of Rev1 was essential for all DRIM events (Supplementary Figures S1 and Supplementary Data; Supplementary Table S2). The rate of GC→CG transversions was reduced >330-fold in the pol3-Y708A rev3Δ strain and 165-fold in the pol3-Y708A rev1Δ strain in comparison with the single pol3-Y708A mutant. The rate of type I complex mutation was reduced >140-fold in both pol3-Y708A rev3Δ and pol3-Y708A rev1Δ strains. Other types of base substitutions and frameshift mutations were also similarly reduced in the rev3Δ and rev1Δ derivatives of pol3-Y708A (Supplementary Figure S1 and Supplementary Table S2). We have additionally tested the effect of the rev1-1 mutation, which results in a Gly193→Arg substitution in the BRCT domain of Rev1 and is thought to affect the structural but not the catalytic function of Rev1 (71). The rev1-1 mutation strongly reduced DRIM (Supplementary Data and Supplementary Table S1), thus providing further support for the importance of the organizing function of Rev1.
Both the structural and the catalytic functions of Rev1 are required for the mutagenic response to replication perturbations. (A) Rate of Canr mutation in the pol3-Y708A strain and its rev1 derivatives. All data are from Supplementary Table S1 and are medians and 95% confidence limits for at least 18 independent cultures. The upper confidence limit is not shown for the strongest mutator strain. The bracket represents the difference in mutation rate for the pol3-Y708A and pol3-Y708A rev1-cd strains. (B) Rates for individual types of base substitutions and complex mutations in the pol3-Y708A and pol3-Y708A rev1-cd strains. Data are from Supplementary Table S2. (C) A working model of complex mutation generation by Pol ζ/Rev1. Base changes of a complex mutation are represented by red asterisks.
Many of the DRIM-associated mutations are base substitutions that could potentially result from a C incorporation [(22); Supplementary Table S2 and Supplementary Figure S1]. Therefore, we asked if the catalytic activity of Rev1 was involved in the generation of some of these mutations. Inactivation of the catalytic activity of Rev1 by a double amino acid change in the active site (D467A, E468A) resulted in a small but significant decrease in DRIM (Figure 1A and Supplementary Table S1). Analysis of the spectrum of Canr mutations in the pol3-Y708A rev1-cd strain revealed that, as expected, the rate of changes to a GC pair (GC→CG, AT→GC, AT→CG), but not to an AT pair (GC→AT, GC→TA, AT→TA), was reduced in strains with the catalytically dead Rev1 (Figure 1B, Supplementary Table S2 and Supplementary Figure S5B). This suggests that an inappropriate C incorporation by Rev1 is responsible for a significant fraction of DRIM-associated base substitutions. The remaining changes to a GC pair likely involve a C or G insertion by Pol ζ or Pol δ itself. Unexpectedly, however, the rate of type I complex mutation was also reduced 7-fold upon inactivation of the deoxycytidyl transferase activity of Rev1 (Figure 1B and Supplementary Table S2). The complex mutations typically require incorporation of various nucleotides, not just a C (22), so the Rev1 deoxycytidyl transferase alone could not possibly generate these mutations. Therefore, it is likely that the DNA polymerase activity responsible for introducing the multiple changes is the one of Pol ζ. The strong reduction in the rate of complex mutations in the pol3-Y708A rev1-cd strain, however, suggests that the C insertion by Rev1 is somehow required for their generation by Pol ζ in vivo. This observation was in apparent disagreement with the fact that purified Pol ζ is perfectly capable of making the complex mutations without the help of Rev1 in vitro (7). In the following sections, we propose and vigorously test a model that resolved this controversy and provided insight into the mechanism of replication-associated Pol ζ- and Rev1-dependent mutagenesis.
A working hypothesis on the mechanism of complex mutation generation by Pol ζ and Rev1
To explain the requirement for the catalytic activity of Rev1 in the formation of complex mutations, we proposed the following model (Figure 1C). DNA polymerases are well known to pause synthesis in a nonrandom fashion during copying of natural DNA templates. We hypothesized that the complex mutations occur at sites where processive synthesis by the replicative polymerase (step a in Figure 1C) pauses at sequences that are for some reasons difficult to copy (step b in Figure 1C). We further propose that Rev1 inserts a ‘C’ at these sites in an attempt to alleviate the stalling (step c in Figure 1C). This is followed by a switch to Pol ζ (step d in Figure 1C). The continuation of DNA synthesis by Pol ζ is accompanied by the generation of a complex mutation (step e in Figure 1C). In this model, the ‘C’ insertion by Rev1 is the initiating event necessary for the creation of a complex mutation by Pol ζ. We proposed that, if the catalytic activity of Rev1 is not available, the stalled replication intermediates (step c) could be processed in an error-free manner by the Rad5/Mms2/Ubc13-dependent pathway. The Mms2-Ubc13 complex and Rad5 are the ubiquitin-conjugating enzyme and the ubiquitin ligase, respectively, that mediate the K63-linked polyubiquitylation of PCNA in response to DNA damage-induced replication blocks. The polyubiquitylation activates a nonmutagenic bypass of DNA lesions that is believed to use an undamaged sister chromatid as a template (72). We proposed that stalled intermediates accumulating in the replication-deficient strain pol3-Y708A are similarly processed via the Rad5/Mms2/Ubc13 pathway if the action of Rev1 deoxycytidyl transferase does not channel them to the mutagenic Pol ζ-dependent pathway (Figure 1C, left). This provides an explanation for the strong reduction in the rate of complex mutations in the rev1-cd derivative of pol3-Y708A.
This hypothesis makes several testable predictions. First, the sites of complex mutations must coincide with sites of the replicative DNA polymerase pausing. Second, Rev1 should be able to extend the primer terminus at the sites where replicative polymerases stall. Third, if neither Rev1 deoxycytidyl transferase nor the Rad5/Mms2/Ubc13 pathway is active, the replication stalling should persist and might provide enough time for Pol ζ to generate the complex mutation on its own. As mentioned earlier, Pol ζ does not require Rev1 to generate complex mutations in vitro when it is given ample time to extend the primer, and the competing error-free bypass is not available (7). Therefore, we predicted that the rate of complex mutation could remain high regardless of the availability of the catalytic activity of Rev1, if the Rad5/Mms2/Ubc13 pathway is inactivated. Experiments described in the subsequent sections test these predictions and, in addition, identify the nature of the ‘difficult’ sequences that cause replication stalling and necessitate the mutagenic bypass by Pol ζ/Rev1.
Sites of the Pol ζ/Rev1-dependent complex mutations coincide with sites of replicative polymerase stalling
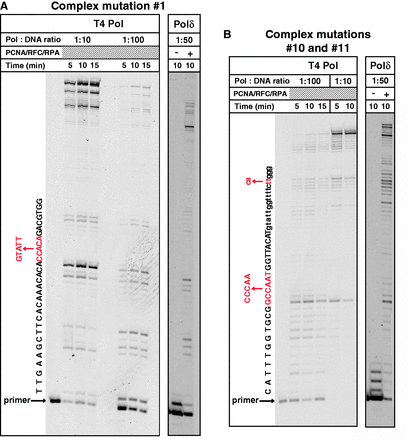
To determine if complex mutations occur at sites of replicative polymerase stalling, we studied the progression of DNA synthesis by T4 DNA polymerase (T4 Pol) and the yeast Pol δ through the yeast CAN1 gene, in which we previously scored the complex mutations in vivo. Single-stranded circular templates for the in vitro reactions were obtained by cloning the CAN1 gene in an M13mp2-based vector in two orientations, such that the pausing pattern during copying of both strands could be analyzed. The complex mutations in the pol3-Y708A strain were observed at a variety of sites within the CAN1 gene (22). We constructed a series of singly primed substrates by annealing the single-stranded M13mp2-CAN1 templates to Cy5-labeled primers complementary to different regions of the CAN1 (Supplementary Figure S2). The location of primers was chosen such that the probability of DNA synthesis termination could be evaluated at each nucleotide position within the first 540 nt of CAN1, a region that spans 11 complex mutation sites (Supplementary Figure S2). Remarkably, for 10 of the 11 sites, a significant impediment of DNA synthesis was seen with both T4 Pol and Pol δ just before the complex mutation site (three examples are shown in Figure 2). The similar pausing pattern observed with T4 Pol and Pol δ suggests that it is the DNA template features and not the polymerase identity that determines the location of pause sites.
Sites of Pol ζ/Rev1-dependent complex mutations coincide with sites of replicative polymerase stalling. DNA synthesis by T4 Pol and yeast Pol δ on singly primed single-stranded M13-CAN1 template was as described in Materials and Methods. Primers A and D (Supplementary Supplementary Data reactions shown in panels (A) and (B), respectively. The sequence of the nascent DNA strand is given to the left of the gel images. The complex mutations observed in the pol3-Y708A strain (22) are shown in red, with numbers indicating nucleotide positions of base substitutions.
The catalytic activity of Rev1 alleviates replicative polymerase stalling at sites of complex mutations
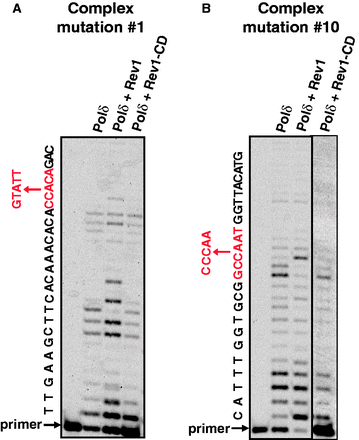
The working model shown in Figure 1C predicts that Rev1 can insert a nucleotide at sites where replicative polymerases pause synthesis. To test this, we repeated the Pol δ primer extension assays described in the previous section in the presence of Rev1 or its catalytically inactive variant. The effect of Rev1 variants on the progression of DNA synthesis through two complex mutation sites in CAN1 is shown in Figure 3. In both cases, Pol δ pauses at a site where the next correct nucleotide to be added is a C. Therefore, Rev1 could potentially take over the primer terminus and incorporate an additional nucleotide if the template sequence at the complex mutation site does not inhibit it as much as it inhibits Pol δ. Indeed, at both complex mutation sites, the presence of the catalytically active Rev1 helped extend the primer by 1 or 2 nt beyond the Pol δ stall sites (Figure 3). In contrast, inclusion of a catalytically inactive Rev1 in the reaction did not alter the Pol δ pausing pattern, indicating that it is the deoxycytidyl transferase activity and not just the physical presence of Rev1 that helped alleviate the stalling. In addition to the known complex mutation sites, the catalytic activity of Rev1 similarly reduced pausing at other sites where Pol δ experienced difficulties extending the primer (see, for example, lower molecular weight bands indicating DNA synthesis pausing ∼10 nt upstream of the complex mutation in Figure 3A).
The catalytic activity of Rev1 alleviates replicative polymerase stalling at sites of complex mutations. DNA synthesis by Pol δ in the presence or absence of Rev1 or Rev1-CD through the sites of complex mutations #1 (A) and #10 (B) was as described in ‘Materials and Methods’ section. All reactions contained PCNA, RFC and RPA. The complex mutation sequences are shown in red to the left of the gel images. The reaction with Pol δ and Rev1-CD shown in the last lane in panel (B) was done in a separate experiment containing 5-fold higher concentrations of PCNA and RFC (0.08 and 0.012 μM, respectively). This was necessary to overcome a strong inhibitory effect of Rev1-CD on Pol δ-dependent synthesis with this substrate, which was presumably owing to efficient binding of Rev1-CD to the primer/template with a G in the first templating position.
In the absence of the Rad5/Mms2/Ubc13 pathway, the catalytic activity of Rev1 is no longer required for DRIM and the generation of complex mutations
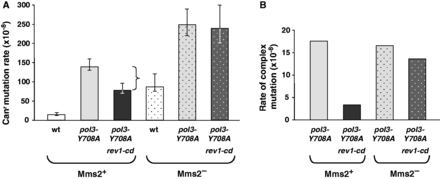
The hypothesis shown in Figure 1C suggests that, in the absence of the catalytic activity of Rev1, the stalled replication intermediates are processed in an error-free manner via the Rad5/Mms2/Ubc13 pathway. This potentially explains the reduced rate of complex mutations in the pol3-Y708A rev1-cd strain in comparison to the pol3-Y708A strain (Figure 1B). As discussed previously, we predicted that inactivation of the error-free pathway could prolong the replication stalling and enable the generation of complex mutations by Pol ζ even if the Rev1 deoxycytidyl transferase is not available. In our experimental system, we expected that the pol3-Y708A rev1-cd double mutants might regain the ability to accumulate complex mutations at a high rate if the Rad5/Mms2/Ubc13 pathway is inactivated. To test this prediction, we compared the rate and spectrum of Canr mutation in the pol3-Y708A and pol3-Y708A rev1-cd strains in the Mms2-deficient background. Consistent with the previously published data (73), the mms2Δ mutation alone results in a mutator phenotype (Figure 4A and Supplementary Table S1), presumably due to channeling the bypass of endogenous DNA lesions to the mutagenic TLS pathway. The rate of Canr mutation in the pol3-Y708A mms2Δ strain was, thus, higher than in the single pol3-Y708A mutant, reflecting the contribution of the mms2Δ mutator. The interaction of the mutator effects of pol3-Y708A and mms2Δ was strictly additive (Figure 4A and Supplementary Table S1), similar to the interaction of pol3-Y708A with other mutations elevating endogenous damage-induced mutagenesis (22). The spectrum of mutations in the pol3-Y708A mms2Δ also appeared to be a combination of the mutational spectra of the single pol3-Y708A and mms2Δ mutants [Supplementary Table S2, Supplementary Figures S5C and Supplementary Data; (22)]. This indicates that the Mms2 defect does not significantly affect the DRIM pathway in strains that carry functional Rev1. The effects of the rev1-cd mutation on DRIM in the Mms2+ and Mms2− strains, however, were dramatically different. In contrast to the Mms-proficient strains and in full agreement with our hypothesis, the rev1-cd mutation did not reduce the overall rate of Canr mutation (Figure 4A and Supplementary Table S1) or the rate of complex mutation (Figure 4B, Supplementary Table S2 and Supplementary Figure S5E) in the Mms2-deficient background. Thus, when the error-free Rad5/Mms2/Ubc13-dependent bypass is inactive, the accumulation of Pol ζ-dependent complex mutations at sites of replication stalling no longer requires the catalytic activity of Rev1.
Effect of the rev1-cd mutation on DRIM in Mms2-proficient and Mms2-deficient background. The diagrams show the rate of Canr mutation (A) and complex mutation (B) in the wild-type, pol3-Y708A and pol3-Y708A rev1-cd strains in the presence and absence of Mms2. Data in (A) are from Supplementary Table S1 and are medians and 95% confidence limits for at least 18 independent cultures. The bracket represents the difference in mutation rate for the pol3-Y708A and pol3-Y708A rev1-cd strains. Data in (B) are from Supplementary Table S2.
The nature of complex mutations reveals template-switching DNA synthesis events triggered by noncanonical DNA structures
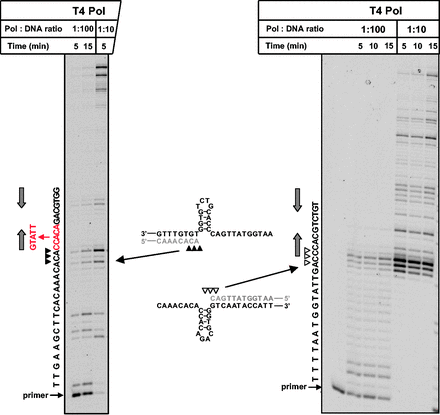
The results described in previous subsections (Figures 2–4) provided strong support for the idea that a large proportion of Pol ζ-dependent mutations occur at sites of replicative polymerase stalling and are triggered by a C incorporation by Rev1 at the beginning of the ‘difficult’ template sequence (Figure 1C). We next asked whether the DNA template sequence at the sites of complex mutations possessed any specific features that caused the replication stalling and required the action of Rev1. It is well known that DNA polymerases can be blocked by non-B DNA structures. Therefore, we analyzed the CAN1 sequence for the presence of repeats that could fold into unusual secondary structures. We found no stable non-B structures resembling those previously implicated in genome instability in prokaryotic or eukaryotic cells (36–49). Remarkably, however, 9 out of 10 polymerase pause sites associated with the Pol ζ-dependent mutations were immediately followed by short (4–6 nt) inverted repeat sequences. This suggested that the polymerase pausing could potentially be caused by small hairpin structures formed by these repeats. To provide additional evidence that these small structures can, in fact, form and inhibit replicative polymerases, we studied the progression of DNA synthesis through a 4-nt repeat sequence in both directions by using either the coding or noncoding strand of CAN1 as a template. With both substrates, the synthesis paused exactly at the base of the proposed hairpin (Figure 5), indicating that the non-B DNA structure was indeed responsible for the pausing. The ability of Rev1 to incorporate a nucleotide after Pol δ has paused (Figure 3), thus, reflects a unique way of handling hairpin template structures by this TLS polymerase.
Stalling occurs at both sides of the hairpins that reside at the sites of Pol ζ/Rev1-dependent complex mutations. The hairpin structures predicted to form at the site of complex mutation # 1 in the coding and noncoding strands of CAN1 are depicted in the middle. The gel images show progression of DNA synthesis by T4 Pol through this region from the left (see also Figure 2A for more detailed analysis of synthesis in this direction) and from the right as indicated by arrows. Primers A and B (Supplementary Figure S2) were used in the reactions shown in the left and right images, respectively. The sequence of the nascent DNA strand is shown on the left of the gel images. The triangles indicate sites of the polymerase stalling. The sequence of complex mutation # 1 is shown in red. Gray arrows show the locations of the inverted repeats that form the hairpin stem.
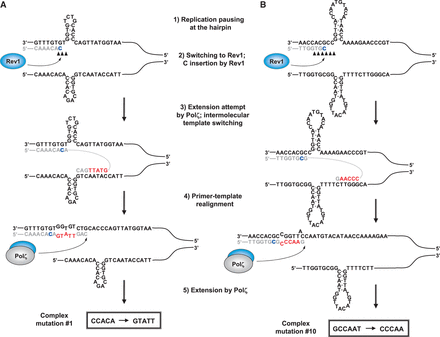
We then hypothesized that the drastic sequence substitutions that we score as complex mutations result not from multiple sequential misincorporations as proposed earlier (7), but rather from template-switching DNA synthesis triggered by replicative polymerase stalling at short-lived hairpin structures. About a quarter of complex mutations observed in the pol3-Y708A strain [Supplementary Figure S3; (22)] involve the introduction of a completely foreign sequence that was long enough to attempt the identification of a region in the vicinity of the small hairpin that could potentially template these events. In all of these mutation cases, the template for the mutant sequence was found on the opposite DNA strand immediately adjacent to the proposed hairpin structure (two examples are shown in Figure 6). The alternative template also contained up to 3 nt following the mutant sequence that, when copied, would allow for the generation of a correctly paired primer terminus after realignment of the nascent strand with the original template. These observations allowed us to propose a more specific model of the complex mutation generation that is supported by our biochemical, genetic and structural analysis (Figure 6). According to this model, replicative polymerases stall synthesis upon encountering short, and presumably transient, hairpin structures (step 1 in Figure 6). Unlike the replicative polymerases, Rev1 is efficient at inserting a nucleotide at the hairpin base, so it takes over the replicative polymerase to extend the primer terminus by 1 or 2 nt (step 2 in Figure 6). A template G near the hairpin base is likely a prerequisite for this step, which was the case for the complex mutation sites that we studied. The incorporation of a C by Rev1 is, thus, not necessarily mutagenic itself. We propose, however, that Rev1 subsequently hands over the primer terminus to Pol ζ, which is followed by an intermolecular template switching (step 3 in Figure 6). The synthesis continues using the opposite strand template until the polymerase approaches the complementary hairpin on the opposite strand. Realignment of the nascent strand with the original correct template (step 4 in Figure 6) produces primer termini that have one to three correctly paired nucleotides at the 3′-end and multiple mismatches further into the double-stranded region. These primer termini might not be good substrates for replicative DNA polymerases that typically require more than three complementary nucleotides at the 3′-end for efficient extension. Pol ζ, in contrast, is highly efficient at extending mismatched primer termini (3). Extension of the ‘realigned’ primer termini by Pol ζ (step 5 in Figure 6) would be consistent with the requirement for this polymerase for the generation of complex mutations in vivo. Not all complex mutation sites we previously observed contain a C at the beginning or before the mutant sequence (22). This is consistent with the observation that the requirement for the catalytic activity of Rev1 in the generation of complex mutations is strong but not absolute (Figure 1B). In contrast, the physical presence of Rev1 was essential for the complex mutation formation: no complex events of any kind were seen in pol3-Y708A rev1Δ strain (Supplementary Table S2 and Supplementary Figure S5A). This may reflect the critical structural role of the Rev1 protein in promoting the template switching, facilitating the extension of multiply mismatched primer terminus by Pol ζ or both. Curiously, complex mutations do occur, albeit rarely, in Pol ζ-deficient strains. They typically involve changes where the original sequence, the mutant sequence or both are >6 nt [(22); Supplementary Figure S3]. In two cases when the mutant sequence was long enough to identify a unique location in the yeast genome where the mutation was templated, the ectopic template was found on a different chromosome (Supplementary Figure S4). Remarkably, this alternative template allowed for synthesis of a DNA stretch that, when realigned to the original CAN1 sequence, creates primer termini with seven or eight correctly paired nucleotides, thus presumably eliminating the requirement for Pol ζ for the extension step.
Template-switching mechanism for the formation of complex mutations. The models in panels (A) and (B) explain the formation of complex mutations # 1 and #10, respectively. Template DNA strands are in black; the correct nascent strand sequences are in gray; the mutant sequences are in red. Black triangles are the replicative polymerase stall sites from Figure 2. Potential sites of C incorporation by Rev1 are in blue (see also Figure 3).
Pol ζ extends mispaired primer termini that we propose are generated during the formation of complex mutations
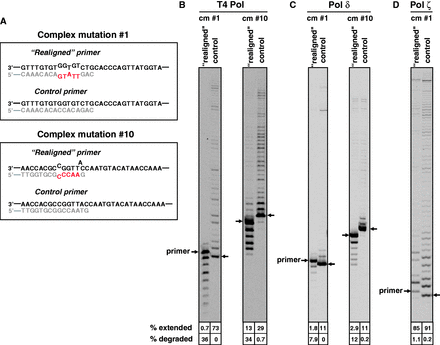
The last step of our model implies that the ‘realigned’ primers containing only one to three correctly paired nucleotides at the 3′-end are extended by Pol ζ to generate the complex mutations. The model also implies that replicative DNA polymerases are not efficient in extending such primer termini because the complex mutations are not formed in the absence of Pol ζ. Therefore, we asked whether the primer termini depicted in Figure 6 could be extended by purified T4 Pol, yeast Pol δ or yeast Pol ζ in vitro. To create singly primed single-stranded DNA substrates for these assays, we used oligonucleotides that generated multiply mismatched primers identical to those shown in Figure 6 (Figure 7A). Fully matched primers that anneal at the same position of the CAN1 were used as a control (Fig. 7A). Both T4 and Pol δ extended the mispaired primer termini extremely inefficiently (Figure 7B and C). Instead, both enzymes degraded these primers using their 3′ exonuclease activity. The control fully matched primers were extended by both T4 and Pol δ, and little or no degradation was observed. In contrast, Pol ζ extended the mismatched and control matched primers with comparable efficiency (Figure 7D), consistent with its proposed role at the last step of complex mutation formation.
Pol ζ extends primers with multiple mismatches that are potentially generated during the formation of complex mutations. (A) Structure of primer-template junctions in the M13-CAN1 substrates used in the experiments in panels (B–D). (B–D) Extension of the ‘realigned’ and the control primers by T4 Pol, yeast Pol δ and Pol ζ was as described in Materials and Methods.
GC→CG transversion hotspots are associated with exceptionally strong DNA polymerase pause sites at a predicted hairpin structure
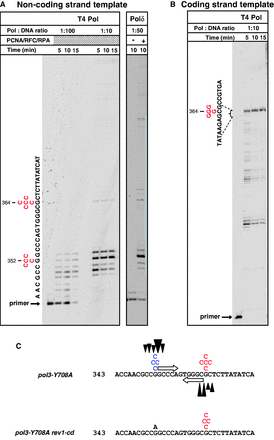
GC→CG transversions constitute the largest class of spontaneous mutations in the pol3-Y708A strain and are entirely Pol ζ-dependent (22). The 540-bp segment of the CAN1 gene, in which we examined DNA polymerase pausing, contains two sites where GC→CG transversions were repeatedly observed (positions 352 and 364; Supplementary Figure S2). Half of all GC→CG substitutions within the 540-bp segment occurred at these hotspots (22). Remarkably, we found that the hotspots immediately flank a DNA region containing 5-nt inverted repeat sequences (Figure 8C). The hairpin formed by these repeats would contain four G•C base pairs in the stem region and, therefore, is predicted to be exceptionally stable. Consistent with this prediction, the strongest DNA synthesis pause sites we observed within the 540-bp region correspond to the base of this hairpin (Figure 8A and B). In fact, very limited, if any, DNA synthesis was seen beyond the hairpin in either direction. Thus, similar to the complex mutations, the GC→CG transversions apparently occur at sites where the replicative polymerase is blocked by a hairpin structure. Because the deoxycydidyl transferase activity of Rev1 can help alleviate the replicative polymerase pausing at such sites (Figure 3), we next considered a possibility that the GC→CG transversions are generated through an erroneous C incorporation by Rev1. Transversions at position 352 would, indeed, be consistent with polymerase pausing during synthesis of the coding strand of CAN1 followed by an incorrect C incorporation (Figure 8C). In contrast, transversions at position 364 would involve pausing during the noncoding strand synthesis and require an incorrect G incorporation, presumably by an enzyme other than Rev1. In excellent agreement with this scenario, GC→CG transversions were no longer observed at position 352 in the rev1-cd strain expressing the catalytically inactive Rev1, while the hotspot at position 364 persisted (Figure 8C and Supplementary Figure S5B). Taken together, the results suggest that GC→CG transversions are strongly promoted by the replicative polymerase stalling at hairpin structures and could occur through incorrect nucleotide incorporation by Rev1 or another DNA polymerase at the hairpin base. Because only 1 nt is altered in these cases, it is not possible to determine whether synthesis on an alternative template is involved in the generation of these mutations. Among nine GC→CG transversions observed previously in the 540-bp CAN1 segment outside the hotspots (22), several map to bases of predicted hairpin structures, while others do not (data not shown). This suggests the existence of additional minor mutational pathways or, potentially, the involvement of less stable DNA secondary structures that our analysis did not identify.
GC→CG transversion hotspots coincide with strong DNA polymerase pause sites. (A) Stalling of T4 Pol and yeast Pol δ at the site of base substitution hotspots at positions 352 and 364 during synthesis of the coding CAN1 strand. (B) Stalling of T4 Pol at the site of the base substitution hotspot at position 364 during synthesis of the noncoding CAN1 strand. The analysis was as in Figure 2 except that primers C and F (Supplementary Figure S2) were used in reactions shown in panels (A) and (B), respectively. (C) Hotspots of Rev1-dependent and Rev1-independent GC→CG transversions are observed at the base of the predicted stable hairpin structure. Sequence of the coding CAN1 strand is shown. Mutations observed at positions 352 and 364 in the pol3-Y708A and pol3-Y708A rev1-cd strains are shown above the sequence of the coding CAN1 strand. Inverted repeats with a strong potential to form a hairpin structure (open arrows) and DNA synthesis pause sites (black triangles) are shown on the top sequence. The pause site above and below the sequence refer to synthesis in the ‘left-to-right’ and ‘right-to-left’ direction, respectively. The size of each triangle approximately corresponds to the severity of the replication block.
DISCUSSION
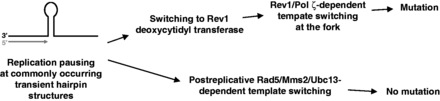
This study reveals the molecular details of the interplay between replicative DNA polymerases, the Rev1 deoxycytidyl transferase, Pol ζ and the Rad5/Mms2/Ubc13 pathway in replication of structurally ambiguous DNA regions. The results suggest that short (4–6 nt) inverted DNA repeats, which were not generally thought to present a challenge for the replication machinery, can fold into hairpin structures and cause replication pausing and activation of mutagenic Pol ζ/Rev1-dependent DNA synthesis. This is consistent with earlier observations that short DNA repeats can form stable hairpins in solution and in double-stranded DNA in vivo (74–77). The stability of the structures depends on the nucleotide composition of the stem and loop regions; stable hairpins with as few as two correctly paired nucleotides in the stem were reported in these studies. The genetic and biochemical data presented here provide strong support for the following model (Figure 9). The replicative polymerase stalling at the base of a small hairpin provides a signal for the recruitment of the TLS polymerase Rev1. The Rev1 is not significantly inhibited by the hairpin and incorporates a nucleotide across from the base of the structure in an attempt to relieve the stalling. The nucleotide incorporation by Rev1 triggers a template-switching event that allows for the hairpin bypass. This bypass is mutagenic owing to the use of an ectopic DNA template and the involvement of the low-fidelity Pol ζ (Figure 9, top). If Rev1 does not take over the primer terminus at the hairpin base, the stalled replication intermediates are processed via the Rad5/Mms2/Ubc13 pathway in an error-free manner (Figure 9, bottom). The availability of Rev1 at replication forks stalled at the hairpin structures may be the key factor determining whether the stalling is resolved via the error-prone or an error-free mechanism.
Pol ζ and Rev1 facilitate the mutagenic bypass of noncanonical secondary structures in DNA.
Previous studies led us to conclude that DRIM does not reflect mutagenic bypass of endogenous DNA lesions but rather results from error-prone copying of undamaged DNA by Pol ζ (22). The identification of sequences with a potential for hairpin structure formation as a major cause of DRIM provides further support to this notion. We observed earlier that combining a DNA replication defect, such as the one caused by the pol3-Y708A mutation, with a nucleotide excision repair or a base excision repair defect causes an additive increase in the mutation rate (22), although all the single mutants are Pol ζ-dependent mutators. The action of Pol ζ on distinct types of substrates, hairpins in the replication-deficient strains and DNA lesions in the repair-deficient strains is in excellent agreement with the observed additive interaction. Similar additivity is seen in this study when the pol3-Y708A and mms2 mutations are combined (Figure 4 and Supplementary Table S1). This is consistent with the view that the mutator phenotype of mms2 strains results from a larger proportion of endogenous DNA lesions being bypassed by the mutagenic Pol ζ-dependent TLS pathway in these strains (73).
The additive interaction of pol3-Y708A and mms2 also indicates that the inactivation of the Rad5/Mms2/Ubc13 pathway does not increase DRIM. In contrast, inactivation of the catalytic activity of Rev1 reduces DRIM, including the template-switching–dependent complex mutations, apparently by channeling the processing of the stalled replication intermediates to the Rad5/Mms2/Ubc13 pathway (Figure 4). This suggests that the Rev1/Pol ζ-dependent and the Rad5/Mms2/Ubc13-dependent bypass pathways do not compete for the same stalled intermediates, but the Rad5/Mms2/Ubc13 pathway rather acts only after the Rev1/Pol ζ-dependent processes are completed. In cells with functional Rev1 and Pol ζ, nearly all replication-blocking structures are processed through the mutagenic pathway, and the error-free pathway serves as a back-up for the cases when Rev1 and Pol ζ fail to bypass the structure. This is consistent with the fact that the mutagenic activity of Rev1 and Pol ζ during DRIM is regulated by monoubiquitylation of PCNA at lysine 164 (6), and the error-free bypass requires polyubiquitylation of PCNA at the same residue (78,79). These processes can only occur sequentially. The Rad5/Mms2/Ubc13 pathway is thought to act postreplicatively, at least in the case of DNA damage bypass (72). A striking feature of the Rev1/Pol ζ-dependent bypass of the proposed hairpin structures is that the ectopic template used for the bypass is found on the opposite strand immediately adjacent to the polymerase stall site (Figure 6). This led us to speculate that, in contrast to the postreplicative error-free bypass, the Rev1/Pol ζ-dependent template switching occurs at the replication fork when the opposite strand is still available in the single-stranded form. It is noteworthy that while the pol3-Y708A strain, like many other strains undergoing DRIM, shows moderate growth deficiency, the rev3, rev1, mms2 or double rev1-cd mms2 mutations do not decrease fitness further (our unpublished observations). This is consistent with the existence of multiple redundant pathways that could resolve stalled replication forks.
Several additional observations suggest that both Rev1 and Pol ζ are needed to keep the template-switching events within close proximity of the original replication stalling site. Although the pol3-Y708A strain could accumulate complex mutations at a high rate in the absence of the catalytic activity of Rev1 if we also inactivated MMS2 (Figure 4, Supplementary Table S2 and Supplementary Figure S3), none of these complex events were templated in the proximity of the mutation site. In these cases, the rather short mutant sequences did not allow us to identify the location of ectopic template sites with any degree of certainty. Analysis of the spectrum of mutations that occurred in the absence of Pol ζ, however, provided an illuminating example. The rare complex mutations found in the pol3-Y708A rev3Δ strain typically involved more extensive sequence changes than those in Pol ζ-proficient strains [(22); Supplementary Figure S3], and the two largest sequence substitutions were clearly templated on a different chromosome (Supplementary Figure S4). Finally, one more confirmation of the role of Pol ζ and Rev1 in limiting the distance at which the template switching can occur comes from the analysis of deletions between short direct repeats in the absence and presence of Pol ζ/Rev1. Such deletions are believed to result from replication slippage (template switching without subsequent realignment of the primer) and are strongly increased by replicative DNA polymerase defects, including the pol3-Y708A (22,80,81). We have shown previously that although inactivation of Pol ζ does not affect the frequency of these events, it greatly increases the size of the deletions, presumably allowing for slippage over greater distances (22). The inactivation of Rev1 had a similar effect on the deletion sizes in the present study (Supplementary Figure S5A). These observations suggest that the biological significance of the Pol ζ/Rev1-dependent template switching could be to provide a means of efficient bypass of replication impediments directly at the fork and prevent primer relocation to distant loci, which could lead to global genomic rearrangements.
The hairpin structure formation in vivo is expected to be inhibited by the single-strand DNA binding protein RPA. Interestingly, we observed similar pausing pattern in the in vitro DNA synthesis reactions with T4 Pol, which were performed in the absence of accessory proteins, and in the yeast Pol δ reactions, which contained enough RPA to cover the entire single-stranded template (Figure 2). Moreover, we observed no difference in the Pol δ stalling pattern when RPA was omitted from the reactions (data not shown). This suggests that RPA may not efficiently prevent the formation of the small hairpin structures.
Pol ζ-dependent complex mutations are also generated during the bypass of endogenous DNA lesions (57,82,83). It is likely that the replicative polymerase stalling is induced by the lesions rather than by noncanonical DNA structures in these cases. To explain the origin of these mutations, the authors proposed a misincorporation-slippage model wherein the mutagenic bypass of a damaged base precedes and initiates a subsequent slippage event. Although DRIM is largely unrelated to the bypass of endogenous lesions (22), we can not exclude that some of the complex mutations we observe occur via the misincorporation-slippage mechanism. Among the complex mutations in the pol3-Y708A strain that involve multiple changes within at least 3 nt, two potentially distinct classes can be noted. About half of the mutations involve a change of a 3–6 nt stretch to a completely different sequence. The other half seemingly involved two different point mutations (base substitutions or frameshifts) separated by two to four unchanged nucleotides (Supplementary Figure S3). We observed that all mutations of the first class (complete sequence replacements) could be easily explained by template switching, using either the opposite strand template as shown in Figure 6 or (in the minority of cases) a nearby ectopic location on the same strand [Supplementary Figure S1 in (22)]. In contrast, only one of four mutations of the second class could be explained by the template-switching mechanism proposed in Figure 6. It is, therefore, possible that at least two distinct pathways exist that lead to complex mutations. Pol ζ, however, is an essential player in each of these pathways, likely because they all require its unique ability to extend primers with multiple mismatched nucleotides.
Complex mutations associated with template switching at short inverted repeat sequences are well known to occur in bacteriophage T4 and E. coli (46,47,52–56). However, the mechanism of these events supported by extensive genetic studies is different from the model for Pol ζ/Rev1-dependent mutagenesis proposed in Figure 6. The Pol ζ/Rev1-dependent mutations in our model are triggered by replicative polymerase stalling at hairpins formed by perfect inverted repeats and involve the use of an alternative template outside of the repeat region. These mutations destroy the original palindromic sequence. In contrast, in the model originally proposed by Lynn Ripley (84) and substantiated by the Lovett laboratory (53–55), complex mutations occur at sites of imperfect inverted repeats when one side of the quasipalindrome is mistakenly used as a template for replication of the other side. The mutations, thus, lead to perfection of the quiasipalindrome. None of the Pol ζ/Rev1-dependent complex mutations we observed could be explained by this mechanism despite the presence of multiple imperfect inverted repeats in the yeast CAN1 gene sequence. This could reflect differences in the severity of replication problems created by perfect and imperfect hairpins in the different experimental systems, the ways the prokaryotic and eukaryotic replication machineries handle hairpin structures, the availability of DNA polymerases capable of template-switching DNA synthesis or other factors.
Several recent reports implicate eukaryotic TLS polymerases in facilitating replication of alternatively structured DNA, including G-quadruplexes and other non-B DNA structures that constitute major obstacles for the replication machinery (85–88). The present study expands these observations by suggesting a role for Pol ζ and Rev1 in the bypass of small hairpin structures that are much more common in all genomes. Furthermore, although it is becoming increasingly clear that DNA sequences capable of adopting non-B DNA conformations are prone to mutation, our study gives this view a new perspective. Under conditions of perturbed replication, we observed little mutagenesis other than that dictated by the local DNA structure. Both complex mutations and base substitution hotspots occurred at sites of replicative polymerase stalling at the predicted small hairpin structures (Figures 2 and 8). In our experimental system, the use of a replication-deficient strain artificially elevated the rate of these events, which allowed us to decipher their mechanism. The significance of these findings, however, is not limited to the specific Pol δ mutant used here. DRIM is caused by a wide variety of defects in the replisome components (6,19,89). Mutations in Pol δ and Pol ε that could potentially result in a defect in the replication fork progression and recruitment of error-prone polymerases have been found in human cancers (90–93). We also previously reported that treatment of wild-type yeast cells with the replication inhibitor hydroxyurea causes a Pol ζ-dependent increase in mutagenesis that resembles DRIM (22). Thus, the results of this study are likely to be applicable to a variety of biologically and clinically important situations. Reduction in the dNTP pool and replication inhibition due to changes in the environment or therapeutic treatments are all expected to trigger Pol ζ- and Rev1-dependent mutagenesis via the mechanism we described.
FUNDING
National Institutes of Health (NIH) [ES015869 to P.V.S., GM032431 to P.M.B.]; NATO [CBP.NR.NRCLG 982734 to P.V.S.]; National Cancer Institute Cancer Biology Training Grant [CA009476 to M.R.N. and T.M.M.]; University of Nebraska Medical Center Graduate Studies Research Fellowship (to T.M.M.). Funding for open access charge: NIH [ES015869 to P.V.S.].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
The authors thank Krista Brown for assistance in preparation of the mutational spectra figures and Kirill Lobachev for helpful discussions.
REFERENCES
Author notes
Present address: Elena I. Stepchenkova, St. Petersburg Branch of the Institute of General Genetics, Universitetskaya emb. 7/9, St. Petersburg 199034, Russia.




Comments