Abstract
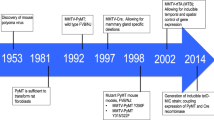
NIH sponsored a meeting of medical and veterinary pathologists with mammary gland expertise in Annapolis in March 1999. Rapid development of mouse mammary models has accentuated the need for definitions of the mammary lesions in genetically engineered mice (GEM) and to assess their usefulness as models of human breast disease. The panel of nine pathologists independently reviewed material representing over 90% of the published systems. The GEM tumors were found to have: (1) phenotypes similar to those of non-GEM; (2) signature phenotypes specific to the transgene; and (3) some morphological similarities to the human disease. The current mouse mammary and human breast tumor classifications describe the majority of GEM lesions but unique morphologic lesions are found in many GEM. Since little information is available on the natural history of GEM lesions, a simple morphologic nomenclature is proposed that allows direct comparisons between models. Future progress requires rigorous application of guidelines covering pathologic examination of the mammary gland and the whole animal. Since the phenotype of the lesions is an essential component of their molecular pathology, funding agencies should adopt policies ensuring careful morphological evaluation of any funded research involving animal models. A pathologist should be part of each research team.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Armes JE, Egan AJ, Southey MC, Dite GS, McCredie MR, Giles GG, Hopper JL and Venter DJ. . 1998 Cancer 83: 2335–2345.
Barnes DM. . 1993 J. Cell. Biochem. Suppl. 17G: 132–138.
Bates P, Fisher R, Ward A, Richardson L, Hill DJ and Graham CF. . 1995 Br. J. Cancer 72: 1189–1193.
Cardiff RD. . 1984 Adv. Cancer Res. 42: 167–190.
Cardiff RD. . 1995 J. Mammary Gland Biology and Neoplasia 1: 61–73.
Cardiff RD. . 1998 J. Mammary Gland Biology and Neoplasia 3: 3–5.
Cardiff RD and Munn RJ. . 1995 Cancer Lett. 90: 13–19.
Cardiff RD and Munn RJ. . 1998 Breast Cancer: Advances in Oncobiology, Vol. 2. Heppner G (ed.).. JAI Press Inc. pp.177–202.
Cardiff RD, Sinn E, Muller W and Leder P. . 1991 Am. J. Pathol. 139: 495–501.
Cardiff RD and Wellings SR. . 1999 J. Mammary Gland Biol. Neoplasia 4: 105–122.
Charpin C, Bacquie N, Bouvier C, Devictor B, Boulat J, Andrac L, Lavaut MN, Allasia C and Piana L. . 1995 Anticancer Res. 15: 2611–2617.
Cheung ATW, Young LJT, Chen PCY, Chao CY, Ndoye A, Barry PA, Muller WJ and Cardiff RD. . 1997 Int. J. Oncology 11: 69–77.
Coffey RJ, Jr., Meise KS, Matsui Y, Hogan BL, Dempsey PJ and Halter SA. . 1994 Cancer Res. 54: 1678–1683.
Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr., Butel JS and Bradley A. . 1992 Nature 356: 215–221.
Dunn TB. . 1959 The physiopathology of cancer, Vol. 2.: F, H (ed.). Hoeber: New York pp.38–84.
Furth PA, BarPeld U, Li MG, Lewis A Laucirica R, Jager R, Weiher H and Russell RG. . 1999 Oncogene 18: 6589–6596.
Gallahan D, Jhappan C, Robinson G, Hennighausen L, Sharp R, Kordon E, Callahan R, Merlino G and Smith GH. . 1996 Cancer Res. 56: 1775–1785.
Genestie C, Zafrani B, Asselain B, Fourquet A, Rozan S, Validire P, Vincent-Salomon A and Sastre-Garau X. . 1998 Anticancer Res. 18: 571–576.
Guy CT, Cardiff RD and Muller WJ. . 1992a Mol. Cell. Biol. 12: 954–961.
Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD and Muller WJ. . 1992b Proc. Natl. Acad. Sci. USA. 89: 10578–10582.
Huseby RA, Soares MJ and Talamantes F. . 1985 Endocrinology 116: 1440–1448.
Husler MR, Kotopoulis KA, Sundberg JP, Tennent BJ, Kunig SV and Knowles BB. . 1998 Transgenic. Res. 7: 253–263.
Iwamoto T, Takahashi M, Ito M, Hamaguchi M, Isobe K, Misawa N, Asai J, Yoshida T and Nakashima I. . 1990 Oncogene 5: 535–542.
Jager R, Herzer U, Schenkel J and Weiher H. . 1997 Oncogene 15: 1787–1795.
Jensen RA, Dupont WD and Page DL. . 1993 J. Cell. Biochem. Suppl. 17G: 59–64.
Jhappan C, Stahle C, Harkins RN, Fausto N, Smith GH and Merlino GT. . 1990 Cell 61: 1137–1146.
Joseph H, Gorska AE, Sohn P, Moses HL and Serra R. . 1999 Mol. Biol. Cell. 10: 1221–1234.
Kordon EC, McKnight RA, Jhappan C, Hennighausen L, Merlino G and Smith GH. . 1995 Dev. Biol. 168: 47–61.
Lakhani SR, Jacquemier J, Sloane JP, Gusterson BA, Anderson TJ, van de Vijver MJ, Farid LM, Venter D, Antoniou A, Storfer-Isser A, Smyth E, Steel CM, Haites N, Scott RJ, Goldgar D, Neuhausen S, Daly PA, Ormiston W, McManus R, Scherneck S, Ponder BA, Ford D, Peto J, Stoppa-Lyonnet D, Easton DF et al. 1998 J. Natl. Cancer. Inst. 90: 1138–1145.
Lane TF and Leder P. . 1997 Oncogene 15: 2133–2144.
Le Doussal V, Tubiana-Hulin M, Friedman S, Hacene K, Spyratos F and Brunet M. . 1989 Cancer 64: 1914–1921.
Li B, Rosen JM, McMenamin-Balano J, Muller WJ and Perkins AS. . 1997 Mol. Cell. Biol. 17: 3155–3163.
Liang TJ, Reid AE, Xavier R, Cardiff RD and Wang TC. . 1996 J. Clin. Invest. 97: 2872–2877.
Kitsberg DI and Leder P. . 1996 Oncogene 13: 2507–2515.
Krane IM and Leder P. . 1996 Oncogene 12: 1781–1788.
Kwan H, Pecenka V, Tsukamoto A, Parslow TG, Guzman R, Lin TP, Muller WJ, Lee FS, Leder P and Varmus HE. . 1992 Mol. Cell. Biol. 12: 147–154.
Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith GH and Callahan R. . 1995 J. Virol. 69: 1932–1938.
Maroulakou IG, Anver M, Garrett L and Green JE. . 1994 Proc. Natl. Acad. Sci. USA. 91: 11236–11240.
Matsui Y, Halter SA, Holt JT, Hogan BL and Coffey RJ. . 1990 Cell 61: 1147–1155.
Medina D. . 1976 Cancer. Res. 36: 2589–2595.
Medina D. . 1988 Carcinogenesis 9: 1113–1119.
Medina D. . 1996 Cancer. Treat. Res. 83: 37–69.
Medina D and Warner MR. . 1976 J. Natl. Cancer. Inst. 57: 331–337.
Morris DW, Barry PA, Bradshaw HD, Jr. and Cardiff RD. . 1990 J. Virol. 64: 1794–1802.
Morris DW and Cardiff RD. . 1987 Adv. in Viral. Oncology 7: pp.123–140.
Muller WJ, Sinn E, Pattengale PK, Wallace R and Leder P. . 1988 Cell 54: 105–115.
Muller WJ, Lee FS, Dickson C, Peters G, Pattengale P and Leder P. . 1990 EMBO J. 9: 907–913.
Page DL, Jensen RA and Simpson JF. . 1998 Breast Cancer Res. Treat. 51: 195–208.
Pravtcheva DD and Wise TL. . 1998 J. Exp. Zool. 281: 43–57.
Rehm S. . 1990 Am. J. Pathol. 136: 575–584.
Rehm S and Liebelt AG. . 1996 Pathobiology of the Aging Mouse, Vol. 2. Mohr U, Dungworth DL, Capen CC, Carlton WW, Sundberg JP and Ward JM. . (eds). ILSI Press: Washington DC pp. 381–398.
Russo J, Gusterson BA, Rogers AE, Russo IH, Wellings SR and van Zwieten MJ. . 1990 Lab. Invest. 62: 244–278.
Sandgren EP, Luetteke NC, Palmiter RD, Brinster RL and Lee DC. . 1990 Cell 61: 1121–1135.
Sandgren EP, Schroeder JA, Qui TH, Palmiter RD, Brinster RL and Lee DC. . 1995 Cancer. Res. 55: 3915–3927.
Sass B and Dunn TB. . 1979 J. Natl. Cancer. Inst. 62: 1287–1293.
Sinn E, Muller W, Pattengale P, Tepler I, Wallace R and Leder P. . 1987 Cell 49: 465–475.
Shibata MA, Maroulakou IG, Jorcyk CL, Gold LG, Ward JM, Green JE. . 1996 Cancer Res 56: 2998–3003.
Smith GH, Gallahan D, Diella F, Jhappan C, Merlino G and Callahan R. . 1995 Cell. Growth. Differ. 6: 563–577.
Stewart TA, Pattengale PK and Leder P. . 1984 Cell 38: 627–637.
Takayama H, LaRochelle WJ, Sharp R, Otsuka T, Kriebel P, Anver M, Aaronson SA and Merlino G. . 1997 Proc. Natl. Acad. Sci. USA. 94: 701–706.
Tavassoli FA. . 1997 The Breast. Journal 3: 48–58.
Tavassoli FA. . 1998 Mod. Pathol. 11: 140–154.
Tavassoli FA and Norris HJ. . 1990 Cancer 65: 518–529.
Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T and Varmus HE. . 1988 Cell 55: 619–625.
Tzeng YJ, Gottlob K, Santarelli R and Graessmann A. . 1996 FEBS. Lett. 380: 215–218.
Tzeng YJ, Guhl E, Graessmann M and Graessmann A. . 1993 Oncogene 8: 1965–1971.
Wang TC, Cardiff RD, Zukerberg L, Lees E, Arnold A and Schmidt EV. . 1994 Nature 369: 669–671.
Wellings SR, Jensen HM and DeVault MR. . 1976 Experientia 32: 1463–1465.
Acknowledgements
The Annapolis meeting was sponsored by the NIH Breast Cancer Think Tank and the NCI Cancer Genome Anatomy Project. The organizers and panelists would like to thank Ms Susan Greenhut for her assistance in organizing the meeting, slides, and logistics for the meetings. We would also like to thank Jai Evans and Ulrike Wagner for their excellent technical assistance throughout the course of this project. The organizers and panelists gratefully acknowledge the following investigators who graciously provided information, blocks and slides to the slide sets: Ari Elson, Priscilla Furth, Jeffrey Green, Hans Weiher, Glenn Merlino, A Graessmann, Izumi Nakashima, Chris Graham, Daniel Medina, Glenn Radice, Eugene Lukanidin, Mark Sternlicht, Barbara Knowles, Dimitrina Pravtcheva, Gilbert Smith, Sonia Jakowlew, Eric Sandgren, Barry Davis, Lothar Hennighausen, Robert Cardiff, Ann Harrington, Jake T Liang, Emmett Schmidt, Philip Leder, Harold Varmus, Robert Coffey, Rob Callahan, Bill Muller, Roy Jensen, Jeffrey Rosen, Hal Moses and Jose Russo. This project has been funded in part with Federal funds from the National Cancer Institute, NIH, under contract no. NO1-CO-56000 and a grant from CGAP.
Author information
Authors and Affiliations
Additional information
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the US Government
IDEALIZED MOUSE WORKUP FOR MAMMARY TUMORIGENESIS STUDIES
IDEALIZED MOUSE WORKUP FOR MAMMARY TUMORIGENESIS STUDIES
The following outline is provided as a guide for maximizing the information obtained from your mouse. Since considerable information can be obtained from analysis of microscopic changes in the mammary gland, it is particularly important to give the pathologist representative ``uninvolved'' mammary tissue in addition to gross mammary lesions. This can give insight into the preneoplastic processes occurring in your model.
PROCEDURE:
1. For any newly generated mouse model, the founder mice and a select number of offspring (ideally no less than 5) should be subjected to a complete necropsy in order to get a complete description of the phenotype. This will allow identification of any additional phenotype changes that might impact on mammary tumorigenesis studies. A guideline for performing complete necropsies can be found at http://www.ncifcrf.gov/vetpath/necropsy.html.
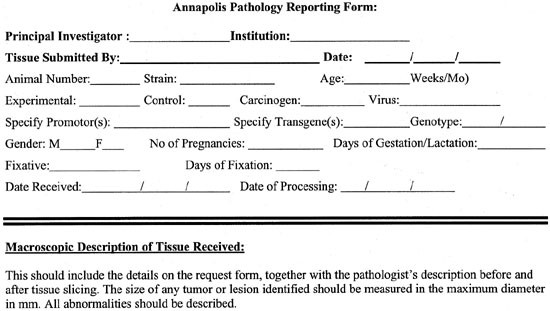
2. For mice in a mammary tumorigenesis study, it is preferable to have a minimum of 10 mice/group. The use of appropriate controls is critical. There should be an equal number of age-matched, non-engineered or untreated controls, of identical genetic background and parity. Note that mammary gland morphology and the incidence of spontaneous mammary tumors varies widely between different genetic backgrounds. This is a particularly important issue when working in a hybrid background. The pathologist should be supplied with the information about the mice found in the Pathology Request Form.
3. For a mammary tumorigenesis study, the mouse should be opened and examined grossly, with any lesions or gross changes noted on the necropsy sheet. Examine all mammary glands grossly and note positions and size of lesions on a diagram of the glands at: URL http://www.ncifcrf.gov/vetpath/necropsy.html. Assign a unique post-mortem number to the mouse.
4. To maximise the amount of information obtainable from each mouse, the tissues indicated in Table 7 should be harvested for each mouse. Tissues of immediate interest should be processed to paraffin block as quickly as possible (within 48h for formalin) as this gives best results for immuno-histochemistry. Organs that are not immediately useful can be fixed and saved for later use (see #8 below). Correct identification of all mice is critical. It is useful to include the identifier (e.g. ears if mice are ear-notched) to allow subsequent cross-checking of mouse ID with PM number.
5. Mammary masses: For masses >0.5cm, it may be desirable to snap freeze half for molecular analysis and fix half for histology (see notes about fixatives below). For smaller masses, fix all for histology. If possible include some uninvolved gland, and harvest the contralateral gland for both histology and molecular analysis.
6. Whole mounts: To visualize the morphology of the ductal tree, it is desirable to whole mount some glands from a select subset of animals. The #4 and #9 (abdominal glands) are generally best for whole mounting because of the presence of the lymph node for orientation. A protocol for mammary whole mounts can be found at http://www-mp.ucdavis.edu/tgmice/Histolab.html. Lesions identified at whole mount can subsequently be sectioned for histology.
7. Trimming and sectioning mammary glands: For trimming mammary glands, remove mammary gland from skin and put on paper (rough DRY brown paper towels work well). Press gland on towel and flatten with forceps. Alternatively, the gland can be spread on a glass slide for better visualization. You can trim off some fat at this point. It is helpful to have a magnifying glass present when trimming muscle away from glands (esp. for thoracic glands). Glands are then fixed (see below).
8. Fixatives:
a. If the study will require use of specific antibodies for immunohistochemistry, it is critical to research the optimal fixative for the antibody of interest in advance. Remember that the mammary epithelium is embedded in excessive amounts of fat that either needs to be trimmed or may require special defatting procedures.
b. For many applications, 10% neutral buffered formalin can be used. Agitate tissues in a 10x-fold excess volume of fixative overnight at room temperature and then change to fresh neutral buffered formalin for storage. For best results with immunohistochemistry, PROCESS TO BLOCK WITHIN 48h. When saving tissues in fixative, double bag all untrimmed tissue with formalin. Recheck bags after 6 mo. and top up with formalin if necessary.
c. For in situ hybridization, best results are usually obtained with 4% paraformaldehyde as fixative. Fix for 3-5d at 4°C.
d. The study and grading of nuclear atypia is better with some acidic or heavy metal fixatives (Bouin's, Zenker's, B-5 ect). Tissue stored in formalin can be post-fixed with these fixatives to improve nuclear detail.
9. Sectioning and staining. When cutting sections from mammary gland blocks, cut longitudinally (“fried egg morphology”. 5u sections are optimal. Hematoxylin/eosin staining is usually optimal for histology. A very light hematoxylin counterstain is generally used alone for immunohistochemistry. However, other nuclear counterstains may be better for specific purposes. Sometimes diagnosis will be aided by additional special stains (e.g. immunohistochemical staining for smooth muscle actin).
Rights and permissions
About this article
Cite this article
Cardiff, R., Anver, M., Gusterson, B. et al. The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting. Oncogene 19, 968–988 (2000). https://doi.org/10.1038/sj.onc.1203277
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1203277
Keywords
This article is cited by
-
Exploring the link between viruses and cancer in companion animals: a comprehensive and comparative analysis
Infectious Agents and Cancer (2023)
-
Molecular Characterization and Landscape of Breast cancer Models from a multi-omics Perspective
Journal of Mammary Gland Biology and Neoplasia (2023)
-
Loss of RPTPγ primes breast tissue for acid extrusion, promotes malignant transformation and results in early tumour recurrence and shortened survival
British Journal of Cancer (2022)
-
Identification of early biomarkers in saliva in genetically engineered mouse model C(3)1-TAg of breast cancer
Scientific Reports (2022)
-
Molecular signatures of in situ to invasive progression for basal-like breast cancers: An integrated mouse model and human DCIS study
npj Breast Cancer (2022)