Key Points
-
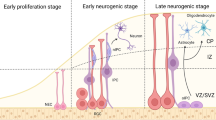
DNA methylation and DNA methyltransferases regulate neurogenesis through epigenetic control of gene expression.
-
Dynamic DNA demethylation exerts temporal control over transcription control during neurogenesis.
-
Cytosine modification derivatives and ten-eleven translocation (TET) proteins precisely coordinate neurogenesis.
-
Histone modification dynamics and their regulators can exert a broad influence over neurogenic processes both directly and indirectly.
Abstract
In the embryonic and adult brain, neural stem cells proliferate and give rise to neurons and glia through highly regulated processes. Epigenetic mechanisms — including DNA and histone modifications, as well as regulation by non-coding RNAs — have pivotal roles in different stages of neurogenesis. Aberrant epigenetic regulation also contributes to the pathogenesis of various brain disorders. Here, we review recent advances in our understanding of epigenetic regulation in neurogenesis and its dysregulation in brain disorders, including discussion of newly identified DNA cytosine modifications. We also briefly cover the emerging field of epitranscriptomics, which involves modifications of mRNAs and long non-coding RNAs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Waddington, C. H. An Introduction to Modern Genetics (G. Allen & Unwin ltd., 1939). This is the first time that the term 'epigenetics' was proposed.
Felsenfeld, G. A brief history of epigenetics. Cold Spring Harbor Persp. Biol. 6, a018200 (2014).
Wu, H. & Zhang, Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156, 45–68 (2014).
Fu, Y., Dominissini, D., Rechavi, G. & He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 15, 293–306 (2014).
Gage, F. H. Mammalian neural stem cells. Science 287, 1433–1438 (2000).
Bond, A. M., Ming, G.-l. & Song, H. Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell 17, 385–395 (2015).
Ming, G. L. & Song, H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70, 687–702 (2011). This review highlights the main principles of adult neurogenesis and summarizes the key remaining questions in the field.
Feliciano, D. M., Bordey, A. & Bonfanti, L. Noncanonical sites of adult neurogenesis in the mammalian brain. Cold Spring Harb. Perspect. Biol. 7, a018846 (2015).
Ma, D. K. et al. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat. Neurosci. 13, 1338–1344 (2010).
Hirabayashi, Y. & Gotoh, Y. Epigenetic control of neural precursor cell fate during development. Nat. Rev. Neurosci. 11, 377–388 (2010).
Gotz, M. & Huttner, W. B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777–788 (2005). This review is a comprehensive summary of the cell biology of embryonic and adult neurogenesis.
Fishell, G. & Kriegstein, A. R. Neurons from radial glia: the consequences of asymmetric inheritance. Curr. Opin. Neurobiol. 13, 34–41 (2003).
Kriegstein, A. R. & Gotz, M. Radial glia diversity: a matter of cell fate. Glia 43, 37–43 (2003).
Noctor, S. C. et al. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J. Neurosci. 22, 3161–3173 (2002).
Hartfuss, E., Galli, R., Heins, N. & Gotz, M. Characterization of CNS precursor subtypes and radial glia. Dev. Biol. 229, 15–30 (2001).
Kessaris, N., Pringle, N. & Richardson, W. D. Specification of CNS glia from neural stem cells in the embryonic neuroepithelium. Phil. Trans. R. Soc. B 363, 71–85 (2008).
Kriegstein, A. & Alvarez-Buylla, A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149–184 (2009). This review systematically summarizes the current state of knowledge of embryonic and adult neurogenesis.
Urban, N. & Guillemot, F. Neurogenesis in the embryonic and adult brain: same regulators, different roles. Front. Cell Neurosci. 8, 396 (2014).
Furutachi, S. et al. Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat. Neurosci. 18, 657–665 (2015).
Fuentealba, L. C. et al. Embryonic origin of postnatal neural stem cells. Cell 161, 1644–1655 (2015).
Sun, G. J. et al. Latent tri-lineage potential of adult hippocampal neural stem cells revealed by Nf1 inactivation. Nat. Neurosci. 18, 1722–1724 (2015).
Alvarez-Buylla, A., Garcia-Verdugo, J. M. & Tramontin, A. D. A unified hypothesis on the lineage of neural stem cells. Nat. Rev. Neurosci. 2, 287–293 (2001).
Cameron, H. A. & McKay, R. D. Adult neurogenesis produces a large pool of new granule cells in the dentate gyrus. J. Comp. Neurol. 435, 406–417 (2001).
Christian, K. M., Song, H. & Ming, G. L. Functions and dysfunctions of adult hippocampal neurogenesis. Annu. Rev. Neurosci. 37, 243–262 (2014).
Zhao, C., Deng, W. & Gage, F. H. Mechanisms and functional implications of adult neurogenesis. Cell 132, 645–660 (2008).
Bird, A. P. CpG-rich islands and the function of DNA methylation. Nature 321, 209–213 (1986). This review highlights the key role of CpG islands and their relation to DNA methylation.
Guo, J. U. et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 14, 1345–1351 (2011). This study is the first systematic analysis of genome-wide CpG methylation changes following neuronal activation in the adult mammalian brain and discusses its potentially unique role in neuronal functions.
Bestor, T. H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 9, 2395–2402 (2000).
Goto, K. et al. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation 56, 39–44 (1994).
Feng, J., Chang, H., Li, E. & Fan, G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 79, 734–746 (2005).
Okano, M., Bell, D. W., Haber, D. A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999).
Li, E., Bestor, T. H. & Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 (1992).
Fan, G. et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development 132, 3345–3356 (2005).
Nguyen, S., Meletis, K., Fu, D., Jhaveri, S. & Jaenisch, R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev. Dyn. 236, 1663–1676 (2007).
Wu, H. et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329, 444–448 (2010). This study uses Dnmt3a -conditional-knockout mice to reveal its critical role in neurogenesis through crosstalk with the repressive histone modification H3K27me3 and its modifiers, the PcG proteins.
Martins-Taylor, K., Schroeder, D. I., LaSalle, J. M., Lalande, M. & Xu, R. H. Role of DNMT3B in the regulation of early neural and neural crest specifiers. Epigenetics 7, 71–82 (2012).
Yoder, J. A., Soman, N. S., Verdine, G. L. & Bestor, T. H. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J. Mol. Biol. 270, 385–395 (1997).
Guo, J. U. et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 17, 215–222 (2014).
Lister, R. et al. Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 (2013). References 38 and 39 describe non-canonical CpH methylation in the mammalian genome.
Gabel, H. W. et al. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 522, 89–93 (2015).
Chen, L. et al. MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc. Natl Acad. Sci. USA 112, 5509–5514 (2015).
Mo, A. et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 (2015).
Zhao, X. et al. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc. Natl Acad. Sci. USA 100, 6777–6782 (2003).
Li, X. et al. Epigenetic regulation of the stem cell mitogen Fgf-2 by Mbd1 in adult neural stem/progenitor cells. J. Biol. Chem. 283, 27644–27652 (2008).
Lewis, J. D. et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69, 905–914 (1992).
Mellen, M., Ayata, P., Dewell, S., Kriaucionis, S. & Heintz, N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430 (2012). This study isolates and profiles the global epigenome and transcriptome of Purkinje cells, granule cells and Bergmann glia from the mouse cerebellum.
Smrt, R. D. et al. Mecp2 deficiency leads to delayed maturation and altered gene expression in hippocampal neurons. Neurobiol. Dis. 27, 77–89 (2007).
Chen, W. G. et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302, 885–889 (2003).
Martinowich, K. et al. DNA methylation-related chromatin remodeling in activity-dependent Bdnf gene regulation. Science 302, 890–893 (2003).
Hu, S. et al. DNA methylation presents distinct binding sites for human transcription factors. eLife 2, e00726 (2013).
Faigle, R. & Song, H. Signaling mechanisms regulating adult neural stem cells and neurogenesis. Biochim. Biophys. Acta 1830, 2435–2448 (2013).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009). This study, for the first time, identifies TET1 as a 2-oxoglutarate (2OG)- and Fe(II)-dependent enzyme that catalyses the conversion of 5mC to 5hmC.
Ito, S. et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 (2010).
He, Y. F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011).
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011). References 54 and 55 together reveal that TET proteins can further convert 5hmC to 5fC and then 5caC, with the latter being converted back to cytosine by thymine DNA glycosylase.
Guo, J. U., Su, Y., Zhong, C., Ming, G. L. & Song, H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434 (2011). This study reveals that TET1 initiates DNA demethylation followed by DNA repair and thus controls key neuronal gene expression.
Ma, D. K. et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077 (2009). This study reveals that DNA demethylation plays a crucial part in controlling key neuronal gene expression.
Rai, K. et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135, 1201–1212 (2008).
Barreto, G. et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 445, 671–675 (2007).
Shin, J., Ming, G. L. & Song, H. Decoding neural transcriptomes and epigenomes via high-throughput sequencing. Nat. Neurosci. 17, 1463–1475 (2014).
Hahn, M. A. et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 3, 291–300 (2013).
Szulwach, K. E. et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 14, 1607–1616 (2011).
Song, C. X. et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 153, 678–691 (2013). This study develops a genome-wide approach to specifically map 5fC-containing DNA.
Yu, M. et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012).
Bachman, M. et al. 5-Formylcytosine can be a stable DNA modification in mammals. Nat. Chem. Biol. 11, 555–557 (2015).
Spruijt, C. G. et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 (2013). This study reveals a range of 5hmC-binding proteins and their differential functions in ES cells and NSCs.
Pichler, G. et al. Cooperative DNA and histone binding by Uhrf2 links the two major repressive epigenetic pathways. J. Cell. Biochem. 112, 2585–2593 (2011).
Huang, Y. et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA 111, 1361–1366 (2014).
Zhang, R. R. et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 13, 237–245 (2013).
Xu, Y. et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell 151, 1200–1213 (2012).
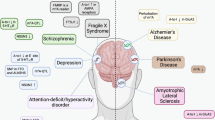
Hsieh, J. & Eisch, A. J. Epigenetics, hippocampal neurogenesis, and neuropsychiatric disorders: unraveling the genome to understand the mind. Neurobiol. Dis. 39, 73–84 (2010).
Wang, F. et al. Genome-wide loss of 5-hmC is a novel epigenetic feature of Huntington's disease. Hum. Mol. Genet. 22, 3641–3653 (2013).
Riccio, A. Dynamic epigenetic regulation in neurons: enzymes, stimuli and signaling pathways. Nat. Neurosci. 13, 1330–1337 (2010).
Thomas, T. & Voss, A. K. Querkopf, a histone acetyltransferase, is essential for embryonic neurogenesis. Front. Biosci. 9, 24–31 (2004).
Greer, E. L. & Shi, Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357 (2012).
Schwartz, Y. B. & Pirrotta, V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8, 9–22 (2007). This is a comprehensive review summarizing the function of Polycomb proteins.
Schuettengruber, B., Martinez, A. M., Iovino, N. & Cavalli, G. Trithorax group proteins: switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 12, 799–814 (2011).
Hirabayashi, Y. et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63, 600–613 (2009).
Pereira, J. D. et al. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl Acad. Sci. USA 107, 15957–15962 (2010).
Fasano, C. A. et al. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell 1, 87–99 (2007).
Lim, D. A. et al. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458, 529–533 (2009).
Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Mohn, F. et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 30, 755–766 (2008).
Sun, G. et al. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol. Cell. Biol. 30, 1997–2005 (2010).
Laurent, B. et al. A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol. Cell 57, 957–970 (2015).
Jepsen, K. et al. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 450, 415–419 (2007).
Yang, X. J. & Seto, E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26, 5310–5318 (2007).
Merson, T. D. et al. The transcriptional coactivator Querkopf controls adult neurogenesis. J. Neurosci. 26, 11359–11370 (2006).
MacDonald, J. L. & Roskams, A. J. Histone deacetylases 1 and 2 are expressed at distinct stages of neuro-glial development. Dev. Dyn. 237, 2256–2267 (2008).
Sun, G., Yu, R. T., Evans, R. M. & Shi, Y. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc. Natl Acad. Sci. USA 104, 15282–15287 (2007).
Gallagher, D. et al. Ankrd11 is a chromatin regulator involved in autism that is essential for neural development. Dev. Cell 32, 31–42 (2015). This study shows that the chromatin modifier ANKRD11 can coordinate with HDAC3 to modulate adult neurogenesis and links its dysregulation to autism spectrum disorders.
Shaked, M. et al. Histone deacetylases control neurogenesis in embryonic brain by inhibition of BMP2/4 signaling. PLoS ONE 3, e2668 (2008).
Hsieh, J., Nakashima, K., Kuwabara, T., Mejia, E. & Gage, F. H. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc. Natl Acad. Sci. USA 101, 16659–16664 (2004).
Xiong, Y. & Guan, K. L. Mechanistic insights into the regulation of metabolic enzymes by acetylation. J. Cell Biol. 198, 155–164 (2012).
Winner, B. & Winkler, J. Adult neurogenesis in neurodegenerative diseases. Cold Spring Harb. Perspect. Biol. 7, a021287 (2015).
Hoglinger, G. U. et al. Dopamine depletion impairs precursor cell proliferation in Parkinson disease. Nat. Neurosci. 7, 726–735 (2004).
Masliah, E. et al. Dopaminergic loss and inclusion body formation in α-synuclein mice: implications for neurodegenerative disorders. Science 287, 1265–1269 (2000).
Nuber, S. et al. Neurodegeneration and motor dysfunction in a conditional model of Parkinson's disease. J. Neurosci. 28, 2471–2484 (2008).
Melrose, H. L. et al. Impaired dopaminergic neurotransmission and microtubule-associated protein tau alterations in human LRRK2 transgenic mice. Neurobiol. Dis. 40, 503–517 (2010).
Chandra, S., Gallardo, G., Fernandez-Chacon, R., Schluter, O. M. & Sudhof, T. C. α-Synuclein cooperates with CSPα in preventing neurodegeneration. Cell 123, 383–396 (2005).
Winner, B. et al. Role of α-synuclein in adult neurogenesis and neuronal maturation in the dentate gyrus. J. Neurosci. 32, 16906–16916 (2012).
Jowaed, A., Schmitt, I., Kaut, O. & Wullner, U. Methylation regulates α-synuclein expression and is decreased in Parkinson's disease patients' brains. J. Neurosci. 30, 6355–6359 (2010).
Matsumoto, L. et al. CpG demethylation enhances alpha-synuclein expression and affects the pathogenesis of Parkinson's disease. PLoS ONE 5, e15522 (2010).
Desplats, P. et al. α-synuclein sequesters Dnmt1 from the nucleus: a novel mechanism for epigenetic alterations in Lewy body diseases. J. Biol. Chem. 286, 9031–9037 (2011).
Winner, B. et al. Adult neurogenesis and neurite outgrowth are impaired in LRRK2 G2019S mice. Neurobiol. Dis. 41, 706–716 (2011).
Beccano-Kelly, D. A. et al. LRRK2 overexpression alters glutamatergic presynaptic plasticity, striatal dopamine tone, postsynaptic signal transduction, motor activity and memory. Hum. Mol. Genet. 24, 1336–1349 (2015).
Cho, H. J. et al. MicroRNA-205 regulates the expression of Parkinson's disease-related leucine-rich repeat kinase 2 protein. Hum. Mol. Genet. 22, 608–620 (2013).
Hardy, J. & Selkoe, D. J. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 297, 353–356 (2002).
Chouraki, V. & Seshadri, S. Genetics of Alzheimer's disease. Adv. Genet. 87, 245–294 (2014).
Kamboh, M. I. Molecular genetics of late-onset Alzheimer's disease. Ann. Hum. Genet. 68, 381–404 (2004).
Games, D. et al. Alzheimer-type neuropathology in transgenic mice overexpressing V717F β-amyloid precursor protein. Nature 373, 523–527 (1995).
Hsiao, K. et al. Correlative memory deficits, Aβ elevation, and amyloid plaques in transgenic mice. Science 274, 99–102 (1996).
Coppieters, N. & Dragunow, M. Epigenetics in Alzheimer's disease: a focus on DNA modifications. Curr. Pharm. Des. 17, 3398–3412 (2011).
Chouliaras, L. et al. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer's disease patients. Neurobiol. Aging 34, 2091–2099 (2013).
De Jager, P. L. et al. Alzheimer's disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat. Neurosci. 17, 1156–1163 (2014).
Handler, M., Yang, X. & Shen, J. Presenilin-1 regulates neuronal differentiation during neurogenesis. Development 127, 2593–2606 (2000).
Bonds, J. A. et al. Presenilin-1 dependent neurogenesis regulates hippocampal learning and memory. PLoS ONE 10, e0131266 (2015).
Kumar, A. & Thakur, M. K. Epigenetic regulation of presenilin 1 and 2 in the cerebral cortex of mice during development. Dev. Neurobiol. 75, 1165–1173 (2015).
Li, G. et al. GABAergic interneuron dysfunction impairs hippocampal neurogenesis in adult apolipoprotein E4 knockin mice. Cell Stem Cell 5, 634–645 (2009).
Yu, C. E. et al. Epigenetic signature and enhancer activity of the human APOE gene. Hum. Mol. Genet. 22, 5036–5047 (2013).
MacDonald, M. E. et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72, 971–983 (1993).
Martindale, D. et al. Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates. Nat. Genet. 18, 150–154 (1998).
Phillips, W., Morton, A. J. & Barker, R. A. Abnormalities of neurogenesis in the R6/2 mouse model of Huntington's disease are attributable to the in vivo microenvironment. J. Neurosci. 25, 11564–11576 (2005).
Lazic, S. E. et al. Decreased hippocampal cell proliferation in R6/1 Huntington's mice. Neuroreport 15, 811–813 (2004).
Kohl, Z. et al. Impaired adult olfactory bulb neurogenesis in the R6/2 mouse model of Huntington's disease. BMC Neurosci. 11, 114 (2010).
Valor, L. M. & Guiretti, D. What's wrong with epigenetics in Huntington's disease? Neuropharmacology 80, 103–114 (2014).
Gorbunova, V., Seluanov, A., Mittelman, D. & Wilson, J. H. Genome-wide demethylation destabilizes CTG·CAG trinucleotide repeats in mammalian cells. Hum. Mol. Genet. 13, 2979–2989 (2004).
Ratovitski, T. et al. Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis. Cell Cycle 11, 2006–2021 (2012).
Shirasaki, D. I. et al. Network organization of the huntingtin proteomic interactome in mammalian brain. Neuron 75, 41–57 (2012).
Ng, C. W. et al. Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proc. Natl Acad. Sci. USA 110, 2354–2359 (2013).
Schoenfeld, T. J. & Cameron, H. A. Adult neurogenesis and mental illness. Neuropsychopharmacology 40, 113–128 (2015).
DeCarolis, N. A. & Eisch, A. J. Hippocampal neurogenesis as a target for the treatment of mental illness: a critical evaluation. Neuropharmacology 58, 884–893 (2010).
Jun, H., Mohammed Qasim Hussaini, S., Rigby, M. J. & Jang, M. H. Functional role of adult hippocampal neurogenesis as a therapeutic strategy for mental disorders. Neural Plast. 2012, 854285 (2012).
Petrik, D., Lagace, D. C. & Eisch, A. J. The neurogenesis hypothesis of affective and anxiety disorders: are we mistaking the scaffolding for the building? Neuropharmacology 62, 21–34 (2012).
Santarelli, L. et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science 301, 805–809 (2003).
Malberg, J. E., Eisch, A. J., Nestler, E. J. & Duman, R. S. Chronic antidepressant treatment increases neurogenesis in adult rat hippocampus. J. Neurosci. 20, 9104–9110 (2000).
Taliaz, D., Stall, N., Dar, D. E. & Zangen, A. Knockdown of brain-derived neurotrophic factor in specific brain sites precipitates behaviors associated with depression and reduces neurogenesis. Mol. Psychiatry 15, 80–92 (2010).
Abuhatzira, L., Makedonski, K., Kaufman, Y., Razin, A. & Shemer, R. MeCP2 deficiency in the brain decreases BDNF levels by REST/CoREST-mediated repression and increases TRKB production. Epigenetics 2, 214–222 (2007).
Klein, M. E. et al. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat. Neurosci. 10, 1513–1514 (2007).
Larimore, J. L. et al. Bdnf overexpression in hippocampal neurons prevents dendritic atrophy caused by Rett-associated MECP2 mutations. Neurobiol. Dis. 34, 199–211 (2009).
Li, W. & Pozzo-Miller, L. BDNF deregulation in Rett syndrome. Neuropharmacology 76, 737–746 (2014).
Pohodich, A. E. & Zoghbi, H. Y. Rett syndrome: disruption of epigenetic control of postnatal neurological functions. Hum. Mol. Genet. 24, R10–R16 (2015).
Zhou, Z. et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron 52, 255–269 (2006).
Li, H. et al. Cell cycle-linked MeCP2 phosphorylation modulates adult neurogenesis involving the Notch signalling pathway. Nat. Commun. 5, 5601 (2014).
Brunet-Gouet, E. & Decety, J. Social brain dysfunctions in schizophrenia: a review of neuroimaging studies. Psychiatry Res. 148, 75–92 (2006).
Reif, A. et al. Neural stem cell proliferation is decreased in schizophrenia, but not in depression. Mol. Psychiatry 11, 514–522 (2006).
Akbarian, S. Epigenetic mechanisms in schizophrenia. Dialogues Clin. Neurosci. 16, 405–417 (2014).
Ikegame, T. et al. DNA methylation of the BDNF gene and its relevance to psychiatric disorders. J. Hum. Genet. 58, 434–438 (2013).
Auta, J. et al. DNA-methylation gene network dysregulation in peripheral blood lymphocytes of schizophrenia patients. Schizophr Res. 150, 312–318 (2013).
Gavin, D. P., Kartan, S., Chase, K., Jayaraman, S. & Sharma, R. P. Histone deacetylase inhibitors and candidate gene expression: an in vivo and in vitro approach to studying chromatin remodeling in a clinical population. J. Psychiatr. Res. 43, 870–876 (2009).
Aberg, K. A. et al. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry 71, 255–264 (2014).
Hannon, E. et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat. Neurosci. 19, 48–54 (2016).
Jaffe, A. E. et al. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat. Neurosci. 19, 40–47 (2016).
Li, X. & Jin, P. Roles of small regulatory RNAs in determining neuronal identity. Nat. Rev. Neurosci. 11, 329–338 (2010).
Clark, B. S. & Blackshaw, S. Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front. Genet. 5, 164 (2014).
Gonzales-Roybal, G. & Lim, D. A. Chromatin-based epigenetics of adult subventricular zone neural stem cells. Front. Genet. 4, 194 (2013).
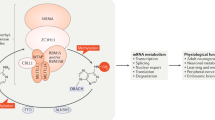
Jia, G., Fu, Y. & He, C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 29, 108–115 (2013).
Yue, Y., Liu, J. & He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355 (2015).
Amiri, A. et al. Pten deletion in adult hippocampal neural stem/progenitor cells causes cellular abnormalities and alters neurogenesis. J. Neurosci. 32, 5880–5890 (2012).
Bian, S. et al. MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep. 3, 1398–1406 (2013).
Dajas-Bailador, F. et al. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat. Neurosci. 15, 697–699 (2012).
Zhao, C., Sun, G., Li, S. & Shi, Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 16, 365–371 (2009).
Sun, G. et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat. Commun. 2, 529 (2011).
Zhao, C. et al. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl Acad. Sci. USA 107, 1876–1881 (2010).
Liu, C. et al. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell 6, 433–444 (2010).
Cheng, L. C., Pastrana, E., Tavazoie, M. & Doetsch, F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408 (2009).
Flynn, R. A. & Chang, H. Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 14, 752–761 (2014).
Bond, A. M. et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12, 1020–1027 (2009).
Ramos, A. D. et al. Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell 12, 616–628 (2013).
Meyer, K. D. & Jaffrey, S. R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15, 313–326 (2014).
Batista, P. J. et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014).
Geula, S. et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015).
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G. & Rottman, F. M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247 (1997).
Bokar, J. A., Rath-Shambaugh, M. E., Ludwiczak, R., Narayan, P. & Rottman, F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 269, 17697–17704 (1994).
Liu, J. et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014).
Ping, X.-L. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189 (2014).
Wang, Y. et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198 (2014).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011). This study, for the first time, identifies fat mass and obesity-associated protein (FTO) as an RNA N6-methyladenosine demethylase.
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Liu, N. et al. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518, 560–564 (2015).
Alarcon, C. R. et al. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 162, 1299–1308 (2015).
Zhou, J. et al. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594 (2015).
Acknowledgements
This work is supported by the US National Institutes of Health (NIH; NS047344 and HD086820 to H.S., NS048271, NS095348, MH110160 and MH105128 to G.L.M., NS051630, NS079625 and MH102690 to P.J.), Dr. Miriam & Sheldon G. Adelson Medical Research Foundation (to G.L.M.) and the Howard Hughes Medical Institute (to C.H.). The authors apologize to colleagues whose work was not cited owing to space limitations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
PowerPoint slides
Glossary
- Neural progenitor cells
-
(NPCs). Precursor cells of the nervous system that can produce more of themselves and differentiate into various types of neural cells.
- Radial glial cells
-
(RGCs). Bipolar cells derived from neuroepithelial cells during embryonic stages that primarily serve as neural progenitor cells during embryonic neurogenesis.
- Imprinted gene silencing
-
A subset of genes that display a parental-specific expression pattern. Compared with normal genes, for which both paternal and maternal alleles are expressed, imprinted genes only express one parental allele. The silencing of one imprinted allele is often mediated by epigenetic mechanisms, such as DNA methylation.
- X-inactivation
-
Females carry two copies of the X chromosome and therefore could potentially express toxic levels (a 'double dose') of X chromosome-linked genes. To prevent this scenario, cells of an early female embryo will randomly inactivate one of the two X chromosomes for gene dosage compensation, termed X-inactivation.
- TET family proteins
-
Ten-eleven translocation (TET) proteins serve as methylcytosine dioxygenases to convert 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine in an iron-dependent manner.
- DNA demethylation
-
An active biochemical process that removes a methyl group from cytosine; this process is catalysed by methylcytosine dioxygenases, such as ten-eleven translocation (TET) proteins.
- Poised enhancers
-
Enhancers refer to the genomic regions that are characterized by uniquely bound transcription factors such as P300 and signature histone modifications such as histone H3 lysine 4 methylation (H3K4me1) that could potentially modulate transcription activation. Poised enhancers bear enhancer characteristics, but their functions are hampered by repressive chromatin marks such that they require additional cues to unleash their functions.
- Transcriptome landscape
-
Global signature transcriptional patterns of different cell types. Maintaining cell type-specific gene expression is crucial for cell identity.
- Interactomes
-
Whole sets of molecules that physically interact with given molecules. In this article, interactome specifically refers to protein–protein interactions.
Rights and permissions
About this article
Cite this article
Yao, B., Christian, K., He, C. et al. Epigenetic mechanisms in neurogenesis. Nat Rev Neurosci 17, 537–549 (2016). https://doi.org/10.1038/nrn.2016.70
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrn.2016.70
This article is cited by
-
Fatty Acid Amide Hydrolase and Cannabinoid Receptor Type 1 Genes Regulation is Modulated by Social Isolation in Rats
Neurochemical Research (2024)
-
Neuroinflammation, memory, and depression: new approaches to hippocampal neurogenesis
Journal of Neuroinflammation (2023)
-
Brain methylome remodeling selectively regulates neuronal activity genes linking to emotional behaviors in mice exposed to maternal immune activation
Nature Communications (2023)
-
In individuals with Williams syndrome, dysregulation of methylation in non-coding regions of neuronal and oligodendrocyte DNA is associated with pathology and cortical development
Molecular Psychiatry (2023)
-
Transcription and DNA methylation signatures of paternal behavior in hippocampal dentate gyrus of prairie voles
Scientific Reports (2023)