Key Points
-
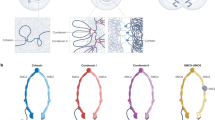
Pericentric heterochromatin is found juxtaposed to the centromere, and is a region of chromatin that remains condensed throughout the cell cycle. Its maintenance is thought to be essential for correct chromosome segregation.
-
At the molecular level, a prominent mark of pericentric heterochromatin is its enrichment in heterochromatin protein-1 (HP1). The multipartite organization of HP1 enables it to bind to numerous nuclear proteins. Among these proteins, those that are potentially involved in the stability of the higher-order structure of pericentric heterochromatin are highlighted. In this way, how HP1 proteins might participate in promoting the self perpetuation of the nuclear domain to which they belong is illustrated.
-
In addition to HP1, epigenetic parameters that are key to the stability of pericentric heterochromatin in mouse are histone-H3 methylated at lysine 9 (H3-K9), hypoacetylated histones, H3-K9 methyltransferase Suv39h and an RNA component. Consistent with this idea, a key aspect in the stability of pericentric heterochromatin is therefore the inter-relationship between these various components. We propose that a local enrichment of HP1 can be achieved by the numerous binding partners that individually have a relatively low affinity.
-
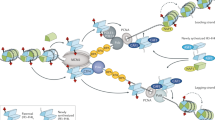
The nature of the interaction of HP1 with its partners within the pericentric heterochromatin domain is highly dynamic, and the properties of HP1 can be integrated into a model for the stable inheritance of the pericentric heterochromatin structure during DNA replication.
-
Finally, exciting recent discoveries implicate RNA in pericentric chromatin maintenance. We compare pericentric heterochromatin in mouse cells and in Schizosaccharomyces pombe and discuss the respective functions of RNA in these regions. An unknown RNA component is required for the maintenance of the mouse pericentric heterochromatin organization, and in S. pombe the RNA-interference machinery is required for the formation of heterochromatin at centromeres.
Abstract
Heterochromatin maintenance is crucial for the clonal inheritance of cell identity, to ensure the proper segregation of chromosomes and the regulation of gene expression. Although it is architecturally stable, heterochromatin has to be flexible to cope with disrupting events such as replication. Recent progress has shed light on the paradoxical properties of heterochromatin in the nucleus, and highlights the roles of heterochromatin protein-1 and, more unexpectedly, RNA molecules in heterochromatin maintenance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Flemming, W. Zell substanz, Kern und Zelltheilung, (Vogel, Leipzig, 1882).
Heitz, E. Das heterochromatin der Moose. Jb. Wiss. Bot. 69, 728 (1928).
Sullivan, B. A., Blower, M. D. & Karpen, G. H. Determining centromere identity: cyclical stories and forking paths. Nature Rev. Genet. 2, 584–596 (2001).
Dillon, N. & Festenstein, R. Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet. 18, 252–258 (2002).
Vissel, B. & Choo, K. H. Mouse major (γ) satellite DNA is highly conserved and organized into extremely long tandem arrays: implications for recombination between nonhomologous chromosomes. Genomics 5, 407–414 (1989).
Taddei, A., Maison, C., Roche, D. & Almouzni, G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nature Cell Biol. 3, 114–120 (2001).
Peters, A. H. et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001).
Ekwall, K., Olsson, T., Turner, B. M., Cranston, G. & Allshire, R. C. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91, 1021–1032 (1997).
Francastel, C., Walters, M. C., Groudine, M. & Martin, D. I. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centromeric heterochromatin. Cell 99, 259–269 (1999).
Brown, K. E., Baxter, J., Graf, D., Merkenschlager, M. & Fisher, A. G. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell 3, 207–217 (1999).
Henikoff, S. Position-effect variegation after 60 years. Trends Genet. 6, 422–426 (1990).
Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002).
Jeppesen, P., Mitchell, A., Turner, B. & Perry, P. Antibodies to defined histone epitopes reveal variations in chromatin conformation and underacetylation of centric heterochromatin in human metaphase chromosomes. Chromosoma 101, 322–332 (1992).
James, T. C. & Elgin, S. C. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 6, 3862–3872 (1986).
James, T. C. et al. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 50, 170–180 (1989).
Eissenberg, J. C. et al. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 87, 9923–9927 (1990).
Singh, P. B. et al. A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res. 19, 789–794 (1991).
Minc, E., Courvalin, J. C. & Buendia, B. HP1γ associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet. Cell Genet. 90, 279–284 (2000).
Nielsen, A. L. et al. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell 7, 729–739 (2001).
Gilbert, N. et al. Formation of facultative heterochromatin in the absence of HP1. Embo J. 22, 5540–5550 (2003).
Jones, D. O., Cowell, I. G. & Singh, P. B. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays 22, 124–137 (2000).
Ball, L. J. et al. Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. Embo J. 16, 2473–2481 (1997).
Nielsen, P. R. et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416, 103–107 (2002).
Brasher, S. V. et al. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 19, 1587–1597 (2000).
Cowieson, N. P., Partridge, J. F., Allshire, R. C. & McLaughlin, P. J. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr. Biol. 10, 517–525 (2000).
Fischle, W. et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17, 1870–1881 (2003).
Li, Y., Kirschmann, D. A. & Wallrath, L. L. Does heterochromatin protein 1 always follow code? Proc. Natl Acad. Sci. USA 99, 16462–16469 (2002).
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001).
Bannister, A. J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Jenuwein, T. & Allis, C. D. Translating the histone code. Science 293, 1074–1080 (2001).
Turner, B. M. Cellular memory and the histone code. Cell 111, 285–291 (2002).
Aagaard, L. et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 18, 1923–1938 (1999).
Grewal, S. I. & Moazed, D. Heterochromatin and epigenetic control of gene expression. Science 301, 798–802 (2003).
Akhtar, A., Zink, D. & Becker, P. B. Chromodomains are protein–RNA interaction modules. Nature 407, 405–409 (2000).
Maison, C. et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nature Genet. 30, 329–334 (2002). This paper shows that a RNA component is required for the maintenance of the mouse pericentric-heterochromatin organization.
Muchardt, C. et al. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep. 3, 975–981 (2002).
Sugimoto, K., Yamada, T., Muro, Y. & Himeno, M. Human homolog of Drosophila heterochromatin-associated protein 1 (HP1) is a DNA-binding protein which possesses a DNA-binding motif with weak similarity to that of human centromere protein C (CENP-C). J. Biochem. (Tokyo) 120, 153–159 (1996).
Meehan, R. R., Kao, C. F. & Pennings, S. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. Embo J. 22, 3164–3174 (2003).
Smothers, J. F. & Henikoff, S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol. 10, 27–30 (2000).
Le Douarin, B. et al. A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. Embo J. 15, 6701–6715 (1996).
Fuks, F., Hurd, P. J., Deplus, R. & Kouzarides, T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 31, 2305–2312 (2003).
Okano, M., Bell, D. W., Haber, D. A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999).
Robertson, K. D. DNA methylation and chromatin — unraveling the tangled web. Oncogene 21, 5361–5379 (2002).
Murzina, N., Verreault, A., Laue, E. & Stillman, B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell 4, 529–540 (1999).
Mello, J. A. & Almouzni, G. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 11, 136–141 (2001).
Nakayama, J., Rice, J. C., Strahl, B. D., Allis, C. D. & Grewal, S. I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 (2001).
Rice, J. C. et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12, 1591–1598 (2003).
Peters, A. H. et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577–1589 (2003).
Jacobs, S. A. & Khorasanizadeh, S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295, 2080–2083 (2002).
Peters, A. H. et al. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nature Genet. 30, 77–80 (2002).
Chadwick, B. P. & Willard, H. F. Chromatin of the Barr body: histone and non-histone proteins associated with or excluded from the inactive X chromosome. Hum. Mol. Genet. 12, 2167–2178 (2003).
Avner, P. & Heard, E. X-chromosome inactivation: counting, choice and initiation. Nature Rev. Genet. 2, 59–67 (2001).
David, G., Turner, G. M., Yao, Y., Protopopov, A. & DePinho, R. A. mSin3-associated protein, mSds3, is essential for pericentric heterochromatin formation and chromosome segregation in mammalian cells. Genes Dev. 17, 2396–2405 (2003).
Rudert, F., Bronner, S., Garnier, J. M. & Dolle, P. Transcripts from opposite strands of γ satellite DNA are differentially expressed during mouse development. Mamm. Genome 6, 76–83 (1995).
Lehnertz, B. et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13, 1192–1200 (2003).
Cheutin, T. et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299, 721–725 (2003).
Festenstein, R. et al. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science 299, 719–721 (2003). References 57 and 58 demonstrate that the heterochromatin protein HP1 is highly dynamic within stable heterochromatin domains.
Gruss, C., Wu, J., Koller, T. & Sogo, J. M. Disruption of the nucleosomes at the replication fork. Embo J. 12, 4533–4545 (1993).
Wolffe, A. P. & Brown, D. D. DNA replication in vitro erases a Xenopus 5S RNA gene transcription complex. Cell 47, 217–227 (1986).
Bozhenok, L., Wade, P. A. & Varga-Weisz, P. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. Embo J. 21, 2231–2241 (2002).
Collins, N. et al. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nature Genet. 32, 627–632 (2002).
McDowell, T. L. et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc. Natl Acad. Sci. USA 96, 13983–13988 (1999).
Nielsen, A. L. et al. Selective interaction between the chromatin-remodeling factor BRG1 and the heterochromatin-associated protein HP1α. Embo J. 21, 5797–5806 (2002).
Taddei, A., Roche, D., Sibarita, J., Turner, B. & Almouzni, G. Duplication and maintenance of heterochromatin domains. J. Cell Biol. 147, 1153–1166 (1999).
Sogo, J. M. & Laskey, R. A. in Chromatin Structure and Gene Expression (ed. Elgin, S. C. R.) 49–71 (Oxford Univ. Press, New York, 1995)
Shibahara, K. I. & Stillman, B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96, 575–585 (1999).
Moggs, J. G. et al. A CAF-1/PCNA mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell. Biol. 20, 1206–1218 (2000).
Sobel, R. E., Cook, R. G., Perry, C. A., Annunziato, A. T. & Allis, C. D. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl Acad. Sci. USA 92, 1237–1241 (1995).
Vermaak, D., Ahmad, K. & Henikoff, S. Maintenance of chromatin states: an open-and-shut case. Curr. Opin. Cell Biol. 15, 266–274 (2003).
Ahmad, K. & Henikoff, S. Epigenetic consequences of nucleosome dynamics. Cell 111, 281–284 (2002).
Ahmad, K. & Henikoff, S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9, 1191–1200 (2002).
Eddy, S. R. Non-coding RNA genes and the modern RNA world. Nature Rev. Genet. 2, 919–929 (2001).
Storz, G. An expanding universe of noncoding RNAs. Science 296, 1260–1263 (2002).
Zamore, P. D. Ancient pathways programmed by small RNAs. Science 296, 1265–1269 (2002).
Finnegan, E. J. & Matzke, M. A. The small RNA world. J. Cell Sci. 116, 4689–4693 (2003).
Park, Y. & Kuroda, M. I. Epigenetic aspects of X-chromosome dosage compensation. Science 293, 1083–1085 (2001).
Matzke, M., Matzke, A. J. & Kooter, J. M. RNA: guiding gene silencing. Science 293, 1080–1083 (2001).
Verheggen, C., Le Panse, S., Almouzni, G. & Hernandez-Verdun, D. Maintenance of nucleolar machineries and pre-rRNAs in remnant nucleolus of erythrocyte nuclei and remodeling in Xenopus egg extracts. Exp. Cell Res. 269, 23–34 (2001).
Volpe, T. A. et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 (2002).
Hall, I. M. et al. Establishment and maintenance of a heterochromatin domain. Science 297, 2232–2237 (2002).
Hall, I. M., Noma, K. & Grewal, S. I. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl Acad. Sci. USA 100, 193–198 (2003).
Volpe, T. et al. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 11, 137–146 (2003). References 81 and 83 demonstrate links between the RNA interference pathway and heterochromatin silencing in fission yeast.
Ramsahoye, B. H. et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA 97, 5237–5242 (2000).
Reinhart, B. J. & Bartel, D. P. Small RNAs correspond to centromere heterochromatic repeats. Science 297, 1831 (2002).
Martienssen, R. A. Maintenance of heterochromatin by RNA interference of tandem repeats. Nature Genet. 35, 213–214 (2003).
Schramke, V. & Allshire, R. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301, 1069–1074 (2003).
Verdel, A. et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303, 672–676 (2004).
Carmell, M. A., Xuan, Z., Zhang, M. Q. & Hannon, G. J. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16, 2733–2742 (2002).
Stein, P., Svoboda, P., Anger, M. & Schultz, R. M. RNAi: mammalian oocytes do it without RNA-dependent RNA polymerase. RNA 9, 187–192 (2003).
Xu, G. L. et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402, 187–191 (1999).
Tamaru, H. & Selker, E. U. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414, 277–283 (2001).
Jackson, J. P., Lindroth, A. M., Cao, X. & Jacobsen, S. E. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416, 556–560 (2002).
Soppe, W. J. et al. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. Embo J. 21, 6549–6559 (2002).
Billy, E., Brondani, V., Zhang, H., Muller, U. & Filipowicz, W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl Acad. Sci. USA 98, 14428–14433 (2001).
Misteli, T. The concept of self-organization in cellular architecture. J. Cell Biol. 155, 181–185 (2001).
Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent on DNA synthesis. Cell 116, 51–61 (2004). This paper describes how histones H3.1 and H3.3 are deposited into chromatin through distinct pathways.
Acknowledgements
We thank E. Heard for discussions and her helpful comments on the review, and R. Martienssen for his comments on the figures. The team of G.A. is supported by grants from Programme Collaboratif Institut Curie/Commissariat à l'Energie Atomique, la Ligue Nationale contre le Cancer (Equipe labellisée la Ligue), Euratom, the Commissariat à l'Energie Atomique and the Research Training Network (RTN).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- EUCHROMATIN
-
A form of chromatin that is de-condensed during interphase.
- KINETOCHORE
-
A multiprotein complex that assembles on centromeric DNA and mediates the attachment and movement of chromosomes along the microtubules of the mitotic spindle.
- CHROMOCENTRE
-
Aggregates of centromere from different chromosomes.
- PERICENTRIC HETEROCHROMATIN
-
A region of chromatin that is found juxtaposed to the centromere and that remains condensed throughout the cell cycle. It is considered to be typical constitutive heterochromatin.
- SATELLITE REPEAT
-
A specific DNA sequence that is repeated many times in a long tandem array.
- POSITION-EFFECT VARIEGATION
-
The heritable suppression of genes that results from their abnormal translocation to a position close to heterochromatin.
- CHROMODOMAIN
-
A highly conserved protein-sequence motif that is common to many chromosomal proteins. In HP1 it is crucial for the interaction with histone H3 that is methylated at lysine 9.
- CHROMOSHADOW DOMAIN
-
A protein-sequence motif that is related to the chromodomain in its amino-acid sequence and has so far only been found in the HP1 family of proteins.
- HISTONE-CODE HYPOTHESIS
-
A hypothesis that proposes that distinct histone modifications, on one or more tails, function sequentially, or in combination, to define a code that is translated by effector proteins with specific biological functions.
- DOSAGE COMPENSATION
-
A process by which the expression of sex-linked genes is equalized in species in which males and females differ in the number of sex chromosomes. In Drosophila melanogaster dosage compensation is achieved by hypertranscription of the single male X chromosome.
- X INACTIVATION
-
A process by which dosage compensation in mammals is achieved by the transcriptional silencing of one of the X chromosomes in XX females.
- RNA INTERFERENCE
-
(RNAi). A post-transcriptional gene-silencing process in which double-stranded RNA triggers the degradation of homologous mRNA.
- ARGONAUTE
-
A family of proteins that are characterized by the presence of two homology domains (PAZ and PIWI) that are essential for the production or function of small interfering RNAs. So far, their biochemical function has not been characterized.
- DICER
-
An RNaseIII-type nuclease that is required for the processing of double-stranded-RNA precursors into small interfering RNAs.
- RNA-DEPENDENT RNA POLYMERASE
-
(Rdp1). An unusual RNA polymerase that uses small interfering RNAs as primers to generate long double-stranded RNA, which in turn can be processed by Dicer.
- SMALL INTERFERING RNA
-
(siRNA). During RNA interference, these non-coding RNAs (around 22 nucleotides long) are derived from the processing of long double-stranded RNA. They direct the destruction of mRNA targets that have the same sequence.
Rights and permissions
About this article
Cite this article
Maison, C., Almouzni, G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 5, 296–305 (2004). https://doi.org/10.1038/nrm1355
Issue Date:
DOI: https://doi.org/10.1038/nrm1355
This article is cited by
-
PRAMEL7 and CUL2 decrease NuRD stability to establish ground-state pluripotency
EMBO Reports (2024)
-
Fluorescent protein lifetimes report densities and phases of nuclear condensates during embryonic stem-cell differentiation
Nature Communications (2023)
-
Diverse silent chromatin states modulate genome compartmentalization and loop extrusion barriers
Nature Structural & Molecular Biology (2023)
-
Regulation of paternal 5mC oxidation and H3K9me2 asymmetry by ERK1/2 in mouse zygotes
Cell & Bioscience (2022)
-
Comparative genomic analysis of the human genome and six bat genomes using unsupervised machine learning: Mb-level CpG and TFBS islands
BMC Genomics (2022)