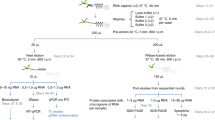
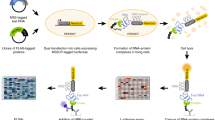
Abstract
Owing to their preeminent biological functions, the repertoire of expressed RNA-binding proteins (RBPs) and their activity states are highly informative about cellular systems. We have developed a novel and unbiased technique, called interactome capture, for identifying the active RBPs of cultured cells. By making use of in vivo UV cross-linking of RBPs to polyadenylated RNAs, covalently bound proteins are captured with oligo(dT) magnetic beads. After stringent washes, the mRNA interactome is determined by quantitative mass spectrometry (MS). The protocol takes 3 working days for analysis of single proteins by western blotting, and about 2 weeks for the determination of complete cellular mRNA interactomes by MS. The most important advantage of interactome capture over other in vitro and in silico approaches is that only RBPs bound to RNA in a physiological environment are identified. When applied to HeLa cells, interactome capture revealed hundreds of novel RBPs. Interactome capture can also be broadly used to compare different biological states, including metabolic stress, cell cycle, differentiation, development or the response to drugs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ascano, M., Hafner, M., Cekan, P., Gerstberger, S. & Tuschl, T. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip Rev. RNA 3, 159–177 (2012).
Konig, J., Zarnack, K., Luscombe, N.M. & Ule, J. Protein-RNA interactions: new genomic technologies and perspectives. Nat. Rev. Genet. 13, 77–83 (2011).
Baltz, A.G. et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 (2012).
Castello, A. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012).
Scherrer, T., Mittal, N., Janga, S.C. & Gerber, A.P. A screen for RNA-binding proteins in yeast indicates dual functions for many enzymes. PLoS ONE 5, e15499 (2010).
Tsvetanova, N.G., Klass, D.M., Salzman, J. & Brown, P.O. Proteome-wide search reveals unexpected RNA-binding proteins in Saccharomyces cerevisiae. PLoS ONE 5, e12671 (2010).
Butter, F., Scheibe, M., Morl, M. & Mann, M. Unbiased RNA-protein interaction screen by quantitative proteomics. Proc. Natl Acad. Sci. USA 106, 10626–10631 (2009).
Anantharaman, V., Koonin, E.V. & Aravind, L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 30, 1427–1464 (2002).
Brimacombe, R., Stiege, W., Kyriatsoulis, A. & Maly, P. Intra-RNA and RNA-protein cross-linking techniques in Escherichia coli ribosomes. Methods Enzymol. 164, 287–309 (1988).
Hockensmith, J.W., Kubasek, W.L., Vorachek, W.R. & von Hippel, P.H. Laser cross-linking of nucleic acids to proteins. Methodology and first applications to the phage T4 DNA replication system. J. Biol. Chem. 261, 3512–3518 (1986).
Hafner, M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010).
Pashev, I.G., Dimitrov, S.I. & Angelov, D. Cross-linking proteins to nucleic acids by ultraviolet laser irradiation. Trends Biochem. Sci. 16, 323–326 (1991).
Suchanek, M., Radzikowska, A. & Thiele, C. Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat. Methods 2, 261–267 (2005).
Creamer, T.J. et al. Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly(A) termination pathway: Nrd1, Nab3, and Sen1. PLoS Genet. 7, e1002329 (2011).
Jamonnak, N. et al. Yeast Nrd1, Nab3, and Sen1 transcriptome-wide binding maps suggest multiple roles in post-transcriptional RNA processing. RNA 17, 2011–2025 (2011).
Jungkamp, A.C. et al. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol. Cell 44, 828–840 (2011).
Hafner, M. et al. PAR-CliP—a method to identify transcriptome-wide the binding sites of RNA binding proteins. J. Vis. Exp. 41, e2034 published online; doi:10.3791/2034 (2010).
Xu, T., Wong, C.C., Kashina, A. & Yates, J.R. Identification of N-terminally arginylated proteins and peptides by mass spectrometry. Nat. Protoc. 4, 325–332 (2009).
Boersema, P.J., Aye, T.T., van Veen, T.A., Heck, A.J. & Mohammed, S. Triplex protein quantification based on stable isotope labeling by peptide dimethylation applied to cell and tissue lysates. Proteomics 8, 4624–4632 (2008).
Choe, L. et al. 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer′s disease. Proteomics 7, 3651–3660 (2007).
Ross, P.L. et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics 3, 1154–1169 (2004).
Ong, S.E. & Mann, M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 (2006).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Wisniewski, J.R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009).
Villen, J. & Gygi, S.P. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 (2008).
Krijgsveld, J., Gauci, S., Dormeyer, W. & Heck, A.J. In-gel isoelectric focusing of peptides as a tool for improved protein identification. J. Proteome Res. 5, 1721–1730 (2006).
Cox, J. et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 (2009).
Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3 published online; doi:10.2202/1544-6115 (2004).
Gentleman, R., Carey, V., Dudoit, S., Irizarry, R. & Huber, W. Bioinformatics and Computational Biology Solutions Using R and Bioconductor (Springer,, 2005).
Acknowledgements
We thank M. Landthaler (Max Delbrück Center for Molecular Medicine) for generously sharing his expertise on PAR-CL. We are grateful to R. Pepperkok (EMBL) for plasmids and M. Gromeier (Duke University Medical Center) for the HeLa Flip-in TRex cell line. We thank M. Muckenthaler (University of Heidelberg) for Huh-7 cells. We acknowledge A. Perez and the EMBL Flow Cytometry Core Facility for FACS experiments, and EMBL Gene and Proteomics Core Facilities for support throughout this work. We also thank Chromotek for expert technical advice and support. A.C. is the beneficiary of a Marie Curie postdoctoral fellowship (FP7). M.W.H. acknowledges support by the European Research Council (ERC) Advanced Grant ERC-2011-ADG_20110310 and the Virtual Liver Network of the German Ministry for Science and Education.
Author information
Authors and Affiliations
Contributions
A.C., J.K., L.M.S. and M.W.H. contributed to the conception and design of the project. A.C., R.H., C.S., B.F. and K.E. carried out the experimental work. A.C. and M.W.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Castello, A., Horos, R., Strein, C. et al. System-wide identification of RNA-binding proteins by interactome capture. Nat Protoc 8, 491–500 (2013). https://doi.org/10.1038/nprot.2013.020
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.020
This article is cited by
-
Control of immediate early gene expression by CPEB4-repressor complex-mediated mRNA degradation
Genome Biology (2022)
-
Alternative polyadenylation by sequential activation of distal and proximal PolyA sites
Nature Structural & Molecular Biology (2022)
-
Comparison of viral RNA–host protein interactomes across pathogenic RNA viruses informs rapid antiviral drug discovery for SARS-CoV-2
Cell Research (2022)
-
ALKBH3 partner ASCC3 mediates P-body formation and selective clearance of MMS-induced 1-methyladenosine and 3-methylcytosine from mRNA
Journal of Translational Medicine (2021)
-
Identification, quantification and bioinformatic analysis of RNA-dependent proteins by RNase treatment and density gradient ultracentrifugation using R-DeeP
Nature Protocols (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.