Abstract
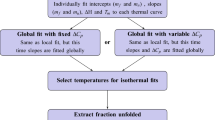
Differential scanning fluorimetry (DSF) is a rapid and inexpensive screening method to identify low-molecular-weight ligands that bind and stabilize purified proteins. The temperature at which a protein unfolds is measured by an increase in the fluorescence of a dye with affinity for hydrophobic parts of the protein, which are exposed as the protein unfolds. A simple fitting procedure allows quick calculation of the transition midpoint; the difference in the temperature of this midpoint in the presence and absence of ligand is related to the binding affinity of the small molecule, which can be a low-molecular-weight compound, a peptide or a nucleic acid. DSF is best performed using a conventional real-time PCR instrument. Ligand solutions from a storage plate are added to a solution of protein and dye, distributed into the wells of the PCR plate and fluorescence intensity measured as the temperature is raised gradually. Results can be obtained in a single day.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Schellman, J.A. Temperature, stability, and the hydrophobic interaction. Biophys. J. 73, 2960–2964 (1997).
Privalov, P.L. Stability of proteins: small globular proteins. Adv. Protein Chem. 33, 167–241 (1979).
Brandts, J.F. Study of strong to ultratight protein interactions using differential scanning calorimetry. Biochemistry 29, 6927–6940 (1990).
Matulis, D., Kranz, J.K., Salemme, F.R. & Todd, M.J. Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry 44, 5258–5266 (2005).
Senisterra, G.A. et al. Screening for ligands using a generic and high-throughput light-scattering-based assay. J. Biomol. Screen. 11, 940–948 (2006).
Vedadi, M. et al. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc. Natl. Acad. Sci. USA 103, 15835–15840 (2006).
Holdgate, G.A. & Ward, W.H. Measurements of binding thermodynamics in drug discovery. Drug Discov. Today 10, 1543–1550 (2005).
Poklar, N., Lah, J., Salobir, M., Macek, P. & Vesnaver, G. pH and temperature-induced molten globule-like denatured states of equinatoxin II: a study by UV-melting, DSC, far- and near-UV CD spectroscopy, and ANS fluorescence. Biochemistry 36, 14345–14352 (1997).
Pantoliano, M.W. et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen. 6, 429–440 (2001).
Lo, M.-C. et al. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal. Biochem. 332, 153–159 (2004).
Ericsson, U.B., Hallberg, M.B., DeTitta, G.T., Dekker, N. & Nordlund, P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 357, 289–298 (2006).
Epps, D.E., Sarver, R.W., Rogers, J.M., Herberg, J.T. & Tomich, P.K. The ligand affinity of proteins measured by isothermal denaturation kinetics. Anal. Biochem. 292, 40–50 (2001).
Plotnikov, V. et al. An autosampling differential scanning calorimeter instrument for studying molecular interactions. Assay Drug Dev. Technol. 1 1 (Part 1): 83–90 (2002).
Pace, C.N., Vajdos, F., Fee, L., Grimsley, G. & Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4, 2411–2423 (1995).
Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 1, 2876–2890 (2006).
Dawson, R.M.C., Elliott, D.C., Elliott, W.H. & Jones, K.M. Data for Biochemical Research 3rd edn. (Oxford University Press, Oxford, UK, 1986).
Acknowledgements
We are grateful to Oleg Y. Fedorov and Guillermo Senisterra for fruitful discussions, and to Aled M. Edwards and Patrick J. Finerty Jr. for critically reading the manuscript. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co. Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research and the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Niesen, F., Berglund, H. & Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2, 2212–2221 (2007). https://doi.org/10.1038/nprot.2007.321
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.321
This article is cited by
-
Exploring the impact of taurine on the biochemical properties of urate oxidase: response surface methodology and molecular dynamics simulation
Journal of Biological Engineering (2024)
-
Rapid evolutionary change in trait correlations of single proteins
Nature Communications (2024)
-
Chloride intracellular channel (CLIC) proteins function as fusogens
Nature Communications (2024)
-
The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition
Communications Biology (2023)
-
ATP induces folding of ALS-causing C71G-hPFN1 and nascent hSOD1
Communications Chemistry (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.