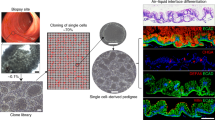
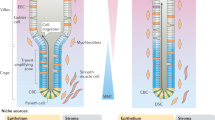
Abstract
Although Lgr5+ intestinal stem cells have been expanded in vitro as organoids, homogeneous culture of these cells has not been possible thus far. Here we show that two small molecules, CHIR99021 and valproic acid, synergistically maintain self-renewal of mouse Lgr5+ intestinal stem cells, resulting in nearly homogeneous cultures. The colony-forming efficiency of cells from these cultures is ∼100-fold greater than that of cells cultured in the absence of CHIR99021 and valproic acid, and multilineage differentiation ability is preserved. We made use of these homogeneous cultures to identify conditions employing simultaneous modulation of Wnt and Notch signaling to direct lineage differentiation into mature enterocytes, goblet cells and Paneth cells. Expansion in these culture conditions may be feasible for Lgr5+ cells from the mouse stomach and colon and from the human small intestine. These methods provide new tools for the study and application of multiple intestinal epithelial cell types.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011).
Yilmaz, Ö.H. et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–495 (2012).
Farin, H.F., van Es, J.H. & Clevers, H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143, 1518–1529 (2012).
Snippert, H.J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Scoville, D.H., Sato, T., He, X.C. & Li, L. Current view: intestinal stem cells and signaling. Gastroenterology 134, 849–864 (2008).
van der Flier, L.G. & Clevers, H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 71, 241–260 (2009).
Crosnier, C., Stamataki, D. & Lewis, J. Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat. Rev. Genet. 7, 349–359 (2006).
Stanger, B.Z., Datar, R., Murtaugh, L.C. & Melton, D.A. Direct regulation of intestinal fate by Notch. Proc. Natl. Acad. Sci. USA 102, 12443–12448 (2005).
Zecchini, V., Domaschenz, R., Winton, D. & Jones, P. Notch signaling regulates the differentiation of post-mitotic intestinal epithelial cells. Genes Dev. 19, 1686–1691 (2005).
Powell, D.W., Pinchuk, I.V., Saada, J.I., Chen, X. & Mifflin, R.C. Mesenchymal cells of the intestinal lamina propria. Annu. Rev. Physiol. 73, 213–237 (2011).
Greenblatt, D.Y. et al. Valproic acid activates Notch-1 signaling and regulates the neuroendocrine phenotype in carcinoid cancer cells. Oncologist 12, 942–951 (2007).
Stockhausen, M.T., Sjölund, J., Manetopoulos, C. & Axelson, H. Effects of the histone deacetylase inhibitor valproic acid on Notch signalling in human neuroblastoma cells. Br. J. Cancer 92, 751–759 (2005).
van der Flier, L.G. et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 136, 903–912 (2009).
de Lau, W. et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 (2011).
Wang, F. et al. Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay. Gastroenterology 145, 383–395 (2013).
van Es, J.H. et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 14, 1099–1104 (2012).
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141, 1762–1772 (2011).
Jung, P. et al. Isolation and in vitro expansion of human colonic stem cells. Nat. Med. 17, 1225–1227 (2011).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Muñoz, J. et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent '+4' cell markers. EMBO J. 31, 3079–3091 (2012).
Yu, D., Cozma, D., Park, A. & Thomas-Tikhonenko, A. Functional validation of genes implicated in lymphomagenesis: an in vivo selection assay using a Myc-induced B-cell tumor. Ann. NY Acad. Sci. 1059, 145–159 (2005).
Milano, J. et al. Modulation of notch processing by γ-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol. Sci. 82, 341–358 (2004).
Wong, G.T. et al. Chronic treatment with the γ-secretase inhibitor LY-411,575 inhibits β-amyloid peptide production and alters lymphopoiesis and intestinal cell differentiation. J. Biol. Chem. 279, 12876–12882 (2004).
van Es, J.H. et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963 (2005).
Fre, S. et al. Notch signals control the fate of immature progenitor cells in the intestine. Nature 435, 964–968 (2005).
Bain, J. et al. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 (2007).
Conti, L. et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 3, e283 (2005).
Pellegrinet, L. et al. Dll1- and Dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 140, 1230–1240 (2011).
Riccio, O. et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 9, 377–383 (2008).
Kazanjian, A., Noah, T., Brown, D., Burkart, J. & Shroyer, N.F. Atonal homolog 1 is required for growth and differentiation effects of notch/gamma-secretase inhibitors on normal and cancerous intestinal epithelial cells. Gastroenterology 139, 918–928 (2010).
Kim, T.H. & Shivdasani, R.A. Genetic evidence that intestinal Notch functions vary regionally and operate through a common mechanism of Math1 repression. J. Biol. Chem. 286, 11427–11433 (2011).
VanDussen, K.L. et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development 139, 488–497 (2012).
D'Souza, B., Miyamoto, A. & Weinmaster, G. The many facets of Notch ligands. Oncogene 27, 5148–5167 (2008).
Shroyer, N.F. et al. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology 132, 2478–2488 (2007).
Han, H. et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14, 637–645 (2002).
Shibata, H. et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science 278, 120–123 (1997).
Barker, N. et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 (2010).
Acknowledgements
This research was supported by US National Institutes of Health (NIH) grant DE013023 to R.L. and a Harvard Institute of Translational Immunology/Helmsley Trust Pilot Grant in Crohn's Disease to J.M.K. H.F.F. was supported by an EMBO long-term fellowship. We also thank D. Breault, R. Montgomery and R. Shivdasani for critically reviewing the manuscript; W. Cho, M. Haraguchi and Q. Wang for helpful discussions; W. Salmon and N. Watson of the W.M. Keck Biological Imaging Facility at the Whitehead Institute for assistance with imaging; G. Paradis, X. Song and M. Jennings of the MIT Flow Cytometry Core Facility for assistance with flow cytometry and CCSG NIH grant CA014051 for support; A. Bhan, V. Yajnik, M. Miri and A. Brunelle at Massachusetts General Hospital for providing human tissue; and all of the staff of the MIT animal care facility.
Author information
Authors and Affiliations
Contributions
X.Y., H.F.F., H.C., R.L. and J.M.K. conceived of and designed the experiments. X.Y. and H.F.F. performed the experiments. J.H.v.E. provided Dll1EGFP-IRES-CreERT2, Atoh1 knockout organoids and Rbpj/Apc knockout-mice intestine. X.Y., H.F.F., J.H.v.E., H.C., R.L. and J.M.K. analyzed the data. X.Y., H.F.F., R.L. and J.M.K. wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Table 1 and Supplementary Results (PDF 4841 kb)
Rights and permissions
About this article
Cite this article
Yin, X., Farin, H., van Es, J. et al. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods 11, 106–112 (2014). https://doi.org/10.1038/nmeth.2737
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2737
This article is cited by
-
Death receptor 5 is required for intestinal stem cell activity during intestinal epithelial renewal at homoeostasis
Cell Death & Disease (2024)
-
Maintenance of adult stem cells from human minor salivary glands via the Wnt signaling pathway
Stem Cell Research & Therapy (2023)
-
A tissue-intrinsic IL-33/EGF circuit promotes epithelial regeneration after intestinal injury
Nature Communications (2023)
-
PhaseFIT: live-organoid phase-fluorescent image transformation via generative AI
Light: Science & Applications (2023)
-
Development of rat duodenal monolayer model with effective barrier function from rat organoids for ADME assay
Scientific Reports (2023)