Abstract
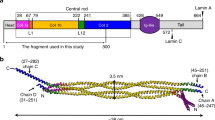
The nuclear lamina is a fundamental constituent of metazoan nuclei. It is composed mainly of lamins, which are intermediate filament proteins that assemble into a filamentous meshwork, bridging the nuclear envelope and chromatin1,2,3,4. Besides providing structural stability to the nucleus5,6, the lamina is involved in many nuclear activities, including chromatin organization, transcription and replication7,8,9,10. However, the structural organization of the nuclear lamina is poorly understood. Here we use cryo-electron tomography to obtain a detailed view of the organization of the lamin meshwork within the lamina. Data analysis of individual lamin filaments resolves a globular-decorated fibre appearance and shows that A- and B-type lamins assemble into tetrameric filaments of 3.5 nm thickness. Thus, lamins exhibit a structure that is remarkably different from the other canonical cytoskeletal elements. Our findings define the architecture of the nuclear lamin meshworks at molecular resolution, providing insights into their role in scaffolding the nuclear lamina.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Aaronson, R. P. & Blobel, G. Isolation of nuclear pore complexes in association with a lamina. Proc. Natl Acad. Sci. USA 72, 1007–1011 (1975)
Burke, B. & Stewart, C. L. The nuclear lamins: flexibility in function. Nature Rev. Mol. Cell Biol. 14, 13–24 (2013)
Fawcett, D. W. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am. J. Anat. 119, 129–145 (1966)
Parry, D. A., Conway, J. F. & Steinert, P. M. Structural studies on lamin. Similarities and differences between lamin and intermediate-filament proteins. Biochem. J. 238, 305–308 (1986)
Dahl, K. N., Engler, A. J., Pajerowski, J. D. & Discher, D. E. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys. J. 89, 2855–2864 (2005)
Lammerding, J. et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 113, 370–378 (2004)
Camozzi, D. et al. Diverse lamin-dependent mechanisms interact to control chromatin dynamics. Focus on laminopathies. Nucleus 5, 427–440 (2014)
Dechat, T., Gesson, K. & Foisner, R. Lamina-independent lamins in the nuclear interior serve important functions. Cold Spring Harb. Symp. Quant. Biol. 75, 533–543 (2010)
Meister, P., Mango, S. E. & Gasser, S. M. Locking the genome: nuclear organization and cell fate. Curr. Opin. Genet. Dev. 21, 167–174 (2011)
Shumaker, D. K., Kuczmarski, E. R. & Goldman, R. D. The nucleoskeleton: lamins and actin are major players in essential nuclear functions. Curr. Opin. Cell Biol. 15, 358–366 (2003)
Aebi, U., Cohn, J., Buhle, L. & Gerace, L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature 323, 560–564 (1986)
Shimi, T. et al. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol. Biol. Cell 26, 4075–4086 (2015)
Harapin, J. et al. Structural analysis of multicellular organisms with cryo-electron tomography. Nature Methods 12, 634–636 (2015)
Mahamid, J. et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972 (2016)
Jones, J. C., Goldman, A. E., Steinert, P. M., Yuspa, S. & Goldman, R. D. Dynamic aspects of the supramolecular organization of intermediate filament networks in cultured epidermal cells. Cell Motil. 2, 197–213 (1982)
Grossman, E. et al. Filaments assembly of ectopically expressed Caenorhabditis elegans lamin within Xenopus oocytes. J. Struct. Biol. 177, 113–118 (2012)
Stuurman, N., Heins, S. & Aebi, U. Nuclear lamins: their structure, assembly, and interactions. J. Struct. Biol. 122, 42–66 (1998)
Heitlinger, E. et al. Expression of chicken lamin B2 in Escherichia coli: characterization of its structure, assembly, and molecular interactions. J. Cell Biol. 113, 485–495 (1991)
Herrmann, H., Bär, H., Kreplak, L., Strelkov, S. V. & Aebi, U. Intermediate filaments: from cell architecture to nanomechanics. Nature Rev. Mol. Cell Biol. 8, 562–573 (2007)
Fisher, D. Z., Chaudhary, N. & Blobel, G. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc. Natl Acad. Sci. USA 83, 6450–6454 (1986)
Dhe-Paganon, S., Werner, E. D., Chi, Y.-I. & Shoelson, S. E. Structure of the globular tail of nuclear lamin. J. Biol. Chem. 277, 17381–17384 (2002)
Ben-Harush, K. et al. The supramolecular organization of the C. elegans nuclear lamin filament. J. Mol. Biol. 386, 1392–1402 (2009)
Zaccai, N. R. et al. A de novo peptide hexamer with a mutable channel. Nature Chem. Biol. 7, 935–941 (2011)
Dittmer, T. A. & Misteli, T. The lamin protein family. Genome Biol. 12, 222 (2011)
Nogales, E., Wolf, S. G., Khan, I. A., Ludueña, R. F. & Downing, K. H. Structure of tubulin at 6.5 A and location of the taxol-binding site. Nature 375, 424–427 (1995)
Galkin, V. E., Orlova, A., Vos, M. R., Schröder, G. F. & Egelman, E. H. Near-atomic resolution for one state of F-actin. Structure 23, 173–182 (2015)
Broers, J. L. V., Ramaekers, F. C. S., Bonne, G., Yaou, R. B. & Hutchison, C. J. Nuclear lamins: laminopathies and their role in premature ageing. Physiol. Rev. 86, 967–1008 (2006)
Worman, H. J. & Courvalin, J.-C. Nuclear envelope, nuclear lamina, and inherited disease. Int. Rev. Cytol. 246, 231–279 (2005)
Worman, H. J., Ostlund, C. & Wang, Y. Diseases of the nuclear envelope. Cold Spring Harb. Perspect. Biol. 2, a000760 (2010)
Fletcher, J. M. et al. A basis set of de novo coiled-coil peptide oligomers for rational protein design and synthetic biology. ACS Synth. Biol. 1, 240–250 (2012)
Guo, M. et al. The role of vimentin intermediate filaments in cortical and cytoplasmic mechanics. Biophys. J. 105, 1562–1568 (2013)
Schirmer, E. C., Guan, T. & Gerace, L. Involvement of the lamin rod domain in heterotypic lamin interactions important for nuclear organization. J. Cell Biol. 153, 479–489 (2001)
Eibauer, M. et al. Unraveling the structure of membrane proteins in situ by transfer function corrected cryo-electron tomography. J. Struct. Biol. 180, 488–496 (2012)
Nickell, S. et al. TOM software toolbox: acquisition and analysis for electron tomography. J. Struct. Biol. 149, 227–234 (2005)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Hotaling, N. A., Bharti, K., Kriel, H. & Simon, C. G. Jr. DiameterJ: a validated open source nanofiber diameter measurement tool. Biomaterials 61, 327–338 (2015)
Trachtenberg, S. & Hammel, I. Determining the persistence length of biopolymers and rod-like macromolecular assemblies from electron microscope images and deriving some of their mechanical properties. Microscopy 1690–1695 (2010)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Wood, C. W. et al. CCBuilder: an interactive web-based tool for building, designing and assessing coiled-coil protein assemblies. Bioinformatics 30, 3029–3035 (2014)
Eibauer, M. et al. Structure and gating of the nuclear pore complex. Nature Commun. 6, 7532 (2015)
Moir, R. D., Donaldson, A. D. & Stewart, M. Expression in Escherichia coli of human lamins A and C: influence of head and tail domains on assembly properties and paracrystal formation. J. Cell Sci. 99, 363–372 (1991)
Acknowledgements
We thank R. Irobalieva for reading the manuscript, Y. Zheng for providing cell lines and L. Gerace for sharing lamin B1 antibody. This work was funded by a Swiss National Science Foundation Grant (SNSF 31003A 159706/1), the Mäxi Foundation and GIF I-1289-412.13/2015 to O.M., and the Forschungskredit of the University of Zurich to Y.T. We thank the Center for Microscopy and Image Analysis at the University of Zurich (ZMB). R.D.G. was funded by National Institutes of Health grant GM106023 and the Progeria Research Foundation. We also thank K. H. Myung for her technical assistance, J. Rappaport of the Nikon Imaging Center in the Feinberg School of Medicine for support, and L. Chang of Nikon Instruments.
Author information
Authors and Affiliations
Contributions
Y.T., R.D.G., K.T.S., M.E. and O.M. conceived and designed the experiments; Y.T. acquired the data and performed and analysed most of the experiments. M.E. analysed the structure of the lamin filaments, and evaluated the lamin gold-label distribution in the immunogold labelling experiments. T.S. and A.E.G. performed the 3D-SIM analysis. M.K. and K.B.H. acquired and analysed the data on the in vitro assembled lamin paracrystals, and A.D.G. refined the structural data within the manuscript revision process. O.M., Y.T., M.E. and K.T.S. discussed analysis, interpretations and presentation. Y.T. and O.M. wrote the manuscript with contributions from all of the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Sample preparation of vimentin deficient MEFs for cryo-ET and 3D-SIM.
a, The 3D-SIM images of immunolabelled wild-type (WT) and Vim−/− MEFs show similar localization and expression of lamin A and lamin B1, derived from n = 3 independent experiments. Scale bar, 5 μm. b, Western blot analyses of the indicated MEF cell lines show unchanged expression levels of each of the lamin isoforms and the retention of the lamins following the short hairpin RNA (shRNA)-mediated knockdown of vimentin. c, The 3D-SIM images of pre-permeabilized, nuclease treated and immunolabelled MEFs show similar localization and expression of lamin A and lamin B1 compared with untreated cells (a). Derived from n = 3 independent experiments. Scale bar, 1 μm. d, The localizations of the lamina-associated proteins (LAPs), emerin and Lap2β exhibit high similarities between pre-permeabilized and nuclease treated (+) and untreated (−) cells, as indicated by 3D-SIM analysis. Derived from n = 3 independent experiments. Scale bar, 1 μm. e, Illustration of the cryo-ET sample preparation procedure.
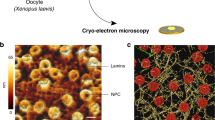
Extended Data Figure 2 Vitrified MEF nuclear envelopes on cryo-ET EM grids.
a, Low-magnification view of two nuclei (encircled by red lines) on a 100 μm × 100 μm carbon mesh of an EM-grid. Scale bar, 50 μm. b, Image shows a part of a nucleus (contoured by the light-grey line). Cryo-tomograms of 1.4 μm × 1.4 μm were acquired at various positions within the area indicated (1–4). Representative xy slices from sub-volumes containing filaments within regions 1–4 are shown in Fig. 1b. Scale bar, 2 μm. c, Higher-magnification projection view of a nucleus reveals some of the canonical components of the nucleus, for example a region of preserved nuclear double-membrane (white arrows), NPCs (white arrowheads) and a highly dense area with putative chromatin remnants (in the upper left corner). Scale bar, 1 μm. n = 55 for a–c. d, Consecutive 1.36 nm xy slices show the thickness of the lamin meshwork and displays occasionally observed putative chromatin remnants (white arrows). Derived from n = 25 sub-volumes. Scale bar, 100 nm. e, Rendering shows segmented lamin filaments (yellow) and putative chromatin remnants (blue). This view of the segmented nuclear lamina displays individual filaments that cross each other at different positions along the z axis within a boundary of ~14 nm. Scale bar, 100 nm.
Extended Data Figure 3 Immunogold labelling of A- and B-type lamins.
Nuclei on EM grids were treated with anti-lamin A/C or anti-lamin B1 antibodies and labelled with 6 nm gold-conjugated protein A before vitrification and cryo-ET analysis. Control samples were treated with protein A conjugate only. a, Projection views of nuclei (contoured by the light-grey line) display the distribution of gold conjugate. Scale bar, 200 nm. b, Zoomed-in images show 9 nm thick xy slices through reconstructed volumes of the respective immunogold-labelled nuclei. A- and B-type filaments are labelled with gold conjugate as indicated (red circles). Scale bar, 100 nm. n = 24 for a and b. c, Gallery view of immunogold-labelled, segmented and skeletonized filaments from 40 nm thick sub-volumes. Green dots indicate immunogold-labelled lamin A and red dots lamin B1 in n = 9 sub-volumes each, locating both within sparsely and densely packed regions. Scale bar, 200 nm. d, Box plot shows the immunogold labelling density of lamin A/C and lamin B1 per μm2 (± s.d.) from volumes shown in c (white line represents the median and black dot the average number of gold particles).
Extended Data Figure 4 Co-immunogold labelling of A- and B-type lamins.
a, Nuclei on EM grids were treated with anti-lamin A/C and anti-lamin B1 and labelled with 6 nm and 10 nm gold conjugates, respectively, before vitrification and cryo-ET analysis, n = 24. Control samples (middle and right) were either treated with anti-lamin A/C before treatment with 6 nm gold conjugate, post-fixation and subsequent treatment with 10 nm gold conjugate (middle), or only with 6 nm and 10 nm gold conjugate (right), omitting incubation with antibodies. Large projection views of parts of nuclei (contoured by the light-grey line) display the distribution of gold conjugates under the indicated conditions. Scale bar, 200 nm. b, Box plots show the labelling density of 6 nm (lamin A/C) and 10 nm (lamin B1) gold particles per μm2 (± s.d.) under the indicated conditions (white line represents the median and black dot the average amount of gold particles). The number of gold particles was extracted from n = 47 sub-volumes (left), n = 9 sub-volumes (middle) or n = 11 sub-volumes (right). c, To validate the reliable assignment of small and large gold colloids, 6 nm and 10 nm gold-conjugated protein A labels were manually picked from samples containing either 6 nm or 10 nm colloids (individual gold colloids), a mixture of both (mixed gold colloids) or from the co-immunogold labelling experiment (lamin A/C and B1 co-labelling experiments) and averaged (n = 200 per condition). The two images at the bottom show a collage of nine individual 6 nm (left) and 10 nm (right) gold-conjugated protein A label, respectively, which were picked from the co-immunogold labelling experiments. The line plot shows the normalized intensity profile of the average diameter of the gold-conjugated protein A labels that were picked from the indicated samples. The solid lines show the intensity profile of the averaged gold label from the samples containing only one type of colloid. The dashed lines show the intensity profile of the averaged gold labels picked from a mixture. The lines with diamonds show the profile of the averaged gold labels from the co-immunogold labelling experiment. The line profiles of the 6 nm and 10 nm averaged gold-conjugated protein A labels are shown in green and red, respectively. The line plot shows almost identical diameters of the 6 nm or 10 nm gold labels for each condition.
Extended Data Figure 5 Lamin filament classification and averaging.
a, Gallery view illustrates a set of extracted and aligned filaments used for further analysis. b, Montage of 36 out of the 40 most populated class averages displays rod-like structures flanked by globular domains at different positions along the central rod. The yellow dots mark four of the most populated classes that are shown in Fig. 3a–c. The class index is given in the upper left corner of the sub-frames and the number of particles in the respective class in the lower left corner. c, Comparison of class averages with index 2 and 5 (Fig. 3c) with in vitro data17 shows remarkable similarities (yellow arrowheads).
Extended Data Figure 6 Structural analysis of in vitro assembled A- and B-type lamin paracrystals.
The final step in the in vitro assembly of assembled A- and B-type lamins results in the formation of paracrystals that display identical organization, corroborating that A- and B-type lamins assemble into similar structures a, TEM analysis and comparison of negatively stained human lamin C (adopted from ref. 41), B1 and B2 paracrystals show an identical striped pattern with 20 nm repeats. n = 20 paracrystals. Scale bar, 20 nm. b, Cryo-ET analysis of in vitro assembled lamin A shows the same 20 nm repeating pattern compared with the lamin isoforms shown in a; n = 9. Scale bar, 20 nm. c, The model shows the 2D arrangement of lamin protofilaments within a paracrystal. The rod-like structure is shown in grey and the globular tail domains in red. d, Averaged structure of in vitro assembled lamin A from cryo-tomograms, as shown in b, displays the striped pattern comprising the purported immunoglobulin-fold domains at distances of 20 nm, comparable to the spacing of the repeating pairs of globular domains in some structural classes from our in situ structural analysis (Fig. 3a, b). The distance of the putative immunoglobulin-fold domains within the stripes of the paracrystals is 6 nm. The averaged structure is derived from n = 4,370 sub-volumes. Scale bar, 20 nm.
Extended Data Figure 7 Lamin assembly scheme.
Averaging and classification of lamin filaments examined in situ show that lamin filaments are composed of two half-staggered head-to-tail polymers. For this, lamin dimers (left) assemble into dimeric head-to-tail polymers exhibiting short overlapping regions, tetrameric in a cross-section (middle). The immunoglobulin domains (red) along the head-to-tail polymer are positioned every ~40 nm. Ultimately, two head-to-tail polymers assemble laterally into a protofilament (right) with a uniformly shaped rod, ~3.5 nm in diameter, containing alternating tetra- and hexameric regions. In this assembly state the immunoglobulin domains are positioned every 20 nm alongside the lamin filament.
Supplementary information
Supplementary Information
This file contains Supplementary Text and References and Supplementary Figure 1. (PDF 477 kb)
Rights and permissions
About this article
Cite this article
Turgay, Y., Eibauer, M., Goldman, A. et al. The molecular architecture of lamins in somatic cells. Nature 543, 261–264 (2017). https://doi.org/10.1038/nature21382
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21382
This article is cited by
-
Simulation and experimental study on the influence of lamina on nanoneedle penetration into the cell nucleus
Biomechanics and Modeling in Mechanobiology (2024)
-
Molecular structure of soluble vimentin tetramers
Scientific Reports (2023)
-
Nuclear lamina strain states revealed by intermolecular force biosensor
Nature Communications (2023)
-
From the membrane to the nucleus: mechanical signals and transcription regulation
Biophysical Reviews (2023)
-
Combined alteration of lamin and nuclear morphology influences the localization of the tumor-associated factor AKTIP
Journal of Experimental & Clinical Cancer Research (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.