Abstract
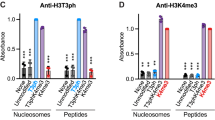
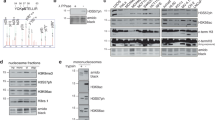
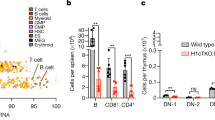
Histones are subject to numerous post-translational modifications1. Some of these ‘epigenetic’ marks recruit proteins that modulate chromatin structure. For example, heterochromatin protein 1 (HP1) binds to histone H3 when its lysine 9 residue has been tri-methylated by the methyltransferase Suv39h (refs 2–6). During mitosis, H3 is also phosphorylated by the kinase Aurora B 7. Although H3 phosphorylation is a hallmark of mitosis, its function remains mysterious. It has been proposed that histone phosphorylation controls the binding of proteins to chromatin8, but any such mechanisms are unknown. Here we show that antibodies against mitotic chromosomal antigens that are associated with human autoimmune diseases9 specifically recognize H3 molecules that are modified by both tri-methylation of lysine 9 and phosphorylation of serine 10 (H3K9me3S10ph). The generation of H3K9me3S10ph depends on Suv39h and Aurora B, and occurs at pericentric heterochromatin during mitosis in different eukaryotes. Most HP1 typically dissociates from chromosomes during mitosis10,11,12, but if phosphorylation of H3 serine 10 is inhibited, HP1 remains chromosome-bound throughout mitosis. H3 phosphorylation by Aurora B is therefore part of a ‘methyl/phos switch’ mechanism8 that displaces HP1 and perhaps other proteins from mitotic heterochromatin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Jenuwein, T. & Allis, C. D. Translating the histone code. Science 293, 1074–1080 (2001)
Bannister, A. J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001)
Jacobs, S. A. et al. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 20, 5232–5241 (2001)
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001)
Nielsen, P. R. et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416, 103–107 (2002)
Jacobs, S. A. & Khorasanizadeh, S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295, 2080–2083 (2002)
Hsu, J. Y. et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279–291 (2000)
Fischle, W., Wang, Y. & Allis, C. D. Binary switches and modification cassettes in histone biology and beyond. Nature 425, 475–479 (2003)
Gitlits, V. M., Macaulay, S. L., Toh, B. H. & Sentry, J. W. Novel human autoantibodies to phosphoepitopes on mitotic chromosomal autoantigens (MCAs). J. Investig. Med. 48, 172–182 (2000)
Murzina, N., Verreault, A., Laue, E. & Stillman, B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell 4, 529–540 (1999)
Sugimoto, K., Tasaka, H. & Dotsu, M. Molecular behaviour in living mitotic cells of human centromere heterochromatin protein HPLα ectopically expressed as a fusion to red fluorescent protein. Cell Struct. Funct. 26, 705–718 (2001)
Schmiedeberg, L., Weisshart, K., Diekmann, S., Meyer Zu Hoerste, G. & Hemmerich, P. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol. Biol. Cell 15, 2819–2833 (2004)
Sullivan, B. A., Blower, M. D. & Karpen, G. H. Determining centromere identity: cyclical stories and forking paths. Nature Rev. Genet. 2, 584–596 (2001)
Mellone, B. G. & Allshire, R. C. Stretching it: putting the CEN(P-A) in centromere. Curr. Opin. Genet. Dev. 13, 191–198 (2003)
Amor, D. J., Kalitsis, P., Sumer, H. & Choo, K. H. Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol. 14, 359–368 (2004)
Fischle, W. et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17, 1870–1881 (2003)
Bernard, P. et al. Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539–2542 (2001)
Nonaka, N. et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nature Cell Biol. 4, 89–93 (2002)
Grewal, S. I. & Rice, J. C. Regulation of heterochromatin by histone methylation and small RNAs. Curr. Opin. Cell Biol. 16, 230–238 (2004)
Earnshaw, W., Bordwell, B., Marino, C. & Rothfield, N. Three human chromosomal autoantigens are recognized by sera from patients with anti-centromere antibodies. J. Clin. Invest. 77, 426–430 (1986)
Hauf, S. et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore–microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161, 281–294 (2003)
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000)
Giet, R. & Glover, D. M. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152, 669–682 (2001)
Mellone, B. G. et al. Centromere silencing and function in fission yeast is governed by the amino terminus of histone H3. Curr. Biol. 13, 1748–1757 (2003)
Wei, Y., Yu, L., Bowen, J., Gorovsky, M. A. & Allis, C. D. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97, 99–109 (1999)
Mateescu, B., England, P., Halgand, F., Yaniv, M. & Muchardt, C. Tethering of HP1 proteins to chromatin is relieved by phosphoacetylation of histone H3. EMBO Rep. 5, 490–496 (2004)
Goto, H., Yasui, Y., Nigg, E. A. & Inagaki, M. Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells 7, 11–17 (2002)
Gimenez-Abian, J. F. et al. Regulation of sister chromatid cohesion between chromosome arms. Curr. Biol. 14, 1187–1193 (2004)
Hirota, T., Gerlich, D., Koch, B., Ellenberg, J. & Peters, J. M. Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 117, 6435–6445 (2004)
Sessa, F. et al. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell 18, 379–391 (2005)
Acknowledgements
We are grateful to K. Mechtler and M. Madalinski for peptide synthesis, to S. Opravil and T. Jenuwein for H3K9me3 antibodies and Suv39h-deficient MEFs, to A. Musacchio for purified Aurora B–INCENP complex, to H. Saya for Aurora A antibodies, and to W. Pollock for collecting MCA sera sent to Gribbles Pathology. T.H. acknowledges a fellowship from the Japanese Society for the Promotion of the Science (JSPS). Research in the laboratory of J.-M.P. is supported by Boehringer Ingelheim, Wiener Wirtschaftsfoerderungsfonds (WWFF), the Sixth Framework Programme of the European Union via the Integrated Project Mitocheck, the Austrian Science Fund and the European Molecular Biology Organization (EMBO). Author Contributions B.-H.T identified MCA sera. T.H. and J.-M.P. conceived and designed the experiments. T.H. and J.J.L. performed the experiments and analysed the data. T.H. and J.-M.P. wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Figure S1
Characterization of MCA2 and MCA6. (JPG 365 kb)
Supplementary Figure S2
The MCA1 epitope is conserved among species. (JPG 243 kb)
Supplementary Figure S3
H3 phosphopeptides do not compete with MCA1 reactivity. (JPG 233 kb)
Supplementary Figure S4
Localization of HP1α in interphase and at different stages of mitosis. (JPG 236 kb)
Supplementary Figure S5
Inhibition of Aurora-B also affects association of HP1β and HP1γ with mitotic chromosomes. (JPG 268 kb)
Supplementary Figure S6
Depletion of Aurora-A and -B. 48 hours after the transfection of indicated siRNAs, cell extracts were analyzed by immunoblotting for Aurora-A and Aurora-B. (JPG 164 kb)
Supplementary Table
Characterization of all six MCA sera. (JPG 272 kb)
Supplementary Figure Legends
Text to accompany the above Supplementary Figures. (DOC 31 kb)
Rights and permissions
About this article
Cite this article
Hirota, T., Lipp, J., Toh, BH. et al. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438, 1176–1180 (2005). https://doi.org/10.1038/nature04254
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature04254
This article is cited by
-
Release of Histone H3K4-reading transcription factors from chromosomes in mitosis is independent of adjacent H3 phosphorylation
Nature Communications (2023)
-
Chromatin-based DNA replication initiation regulation in eukaryotes
Genome Instability & Disease (2023)
-
Linking DNA repair and cell cycle progression through serine ADP-ribosylation of histones
Nature Communications (2022)
-
A Mediator-cohesin axis controls heterochromatin domain formation
Nature Communications (2022)
-
Role of aurora kinase B in regulating resistance to paclitaxel in breast cancer cells
Human Cell (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.