Abstract
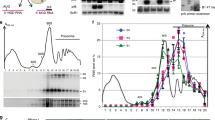
The selective degradation of messenger RNAs enables cells to regulate the levels of particular mRNAs in response to changes in the environment. Ribonuclease (RNase) E (ref. 1), a single-strand-specific endonuclease2,3,4 that is found in a multi-enzyme complex known as the ‘degradosome’5,6,7, initiates the degradation of many mRNAs in Escherichia coli3,8,9. Its relative lack of sequence specificity and the presence of many potential cleavage sites in mRNA substrates2,3 cannot explain why mRNA decay frequently proceeds in a net 5′-to-3′ direction9,10,11. I have prepared covalently closed circular derivatives of natural substrates, the rpsT mRNA encoding ribosomal protein S20 (ref. 2) and the 9S precursor to 5S ribosomal RNA1,12, and find that these derivatives are considerably more resistant to cleavage in vitro by RNase E than are linear molecules. Moreover, antisense oligo-deoxynucleotides complementary to the 5′ end of linear substrates significantly reduce the latter's susceptibility to attack by RNase E. Finally, natural substrates with terminal 5′-triphosphate groups are poorly cleaved by RNase E in vitro, whereas 5′ monophosphorylated substrates are strongly preferred (compare with ref. 13). These results show that RNase E has inherent vectorial properties, with its activity depending on the 5′ end of its substrates; this can account for the direction of mRNA decay in E. coli, the phenomenon of ‘all or none’ mRNA decay, and the stabilization provided by 5′ stem–loop structures14,15,16,17.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Misra, T. K. & Apirion, D. RNase E, an RNA processing enzyme from Escherichia coli. J. Biol. Chem. 254, 11154–11159 (1979).
Mackie, G. A. Secondary structure of the mRNA for ribosomal protein S20. J. Biol. Chem. 267, 1054–1061 (1992).
Ehretsmann, C. P., Carpousis, A. J. & Krisch, H. M. Specificity of Escherichia coli endoribonuclease RNase E: in vivo and in vitro analysis of mutants in a bacteriophage T4 mRNA processing site. Genes Dev. 6, 149–159 (1992).
McDowall, K. J., Lin-Chao, S. & Cohen, S. N. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J. Biol. Chem. 269, 10790–10796 (1994).
Carpousis, A. J., Van Houwe, G., Ehretsmann, C. & Krisch, H. Copurification of E. coli RNase E and PNPase: evidence for a specific association between two enzymes important in mRNA processing and degradation. Cell 76, 889–900 (1994).
Miczak, A., Kaberdin, V. R., Wei, C.-L. & Lin-Chao, S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl Acad. Sci. USA 93, 3865–3869 (1996).
Py, B., Higgins, C. F., Krisch, H. & Carpousis, A. J. ADEAD-box RNA helicase in the Escherichia coli degradosome. Nature 381, 169–172 (1996).
Melefors, Ö Lundberg, U. & von Gabain, A. in Control of Messenger RNA Stability (eds Belasco, J.& Brawerman, G.) 53–70 (Academic, San Diego, (1993)).
Nierlich, D. P. & Murakawa, G. J. The decay of bacterial messenger RNA. Progr. Nucleic Acid Res. Mol. Biol. 52, 153–216 (1996).
Apirion, D. Degradation of RNA in Escherichia coli: a hypothesis. Mol. Gen. Genet. 122, 313–322 (1972).
Kennell, D. in Maximising Gene Expression (eds Reznikoff, W. S.& Gold, L.) 101–142 (Butterworths, Stoneham, MA, (1986)).
Cormack, R. S. & Mackie, G. A. Structural requirements for the processing of Escherichia coli 5S ribosomal RNA by RNase E in vitro. J. Mol. Biol. 228, 1078–1090 (1992).
Lin-Chao, S. & Cohen, S. N. The rate of processing and degradation of antisense RNA1 regulates the replication of ColE1-type plasmids in vivo. Cell 65, 1233–1242 (1991).
Emory, S. A., Bouvet, P. & Belasco, J. G. A5′ terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 6, 135–148 (1992).
Bouvet, P. & Belasco, J. G. Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature 360, 488–491 (1992).
Hansen, M. J. et al . The ompA 5′ untranslated region impedes a major pathway for mRNA degradation in Escherichia coli. Mol. Microbiol. 12, 707–716 (1994).
Mackie, G. A., Genereaux, J. G. & Masterman, S. K. Modulation of the activity of RNase E in vitro by RNA sequences and secondary structures 5′ to cleavage sites. J. Biol. Chem. 272, 609–616 (1997).
Moore, M. J. & Sharp, P. A. Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at splice sites. Science 256, 992–997 (1992).
Coburn, G. A. & Mackie, G. A. Reconstitution of the degradation of the mRNA for ribosomal protein S20 with purified enzymes. J. Mol. Biol. 279, 1061–1074 (1998).
Cormack, R. S., Genereaux, J. G. & Mackie, G. A. RNase E activity is conferred by a single polypeptide: overexpression, purification, and properties of the ams/rne/hmp1 gene product. Proc. Natl Acad. Sci. USA 90, 9006–9010 (1993).
Mackie, G. A. Specific endonucleolytic cleavage of the mRNA for ribosomal protein S20 of Escherichia coli requires the product of the ams gene in vivo and in vitro. J. Bacteriol. 173, 2488–2497 (1991).
Huang, H., Liao, J. & Cohen, S. N. Poly(A)- and poly(U)-specific RNA 3′ tail shortening by E. coli ribonuclease E. Nature 391, 99–102 (1998).
Xu, F. & Cohen, S. N. RNA degradation in Escherichia coli regulated by 3′ adenylation and 5′ phosphorylation. Nature 374, 180–183 (1995).
Beelman, C. A. et al . An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382, 642–646 (1996).
Couttet, P. et al . Messenger RNA deadenylation precedes decapping in mammalian cells. Proc. Natl Acad. Sci. USA 94, 5628–5633 (1997).
Zhu, L., Gangopadhyay, T., Padmanabda, K. P. & Deutscher, M. P. Escherichia coli rna gene encoding RNase I: cloning, overexpression, subcellular distribution of the enzyme and use of an rna deletion to identify additional RNases. J. Bacteriol. 172, 3146–3151 (1990).
Mackie, G. A. & Genereaux, J. G. The role of RNA structure in determining RNase E-dependent cleavage sites in the mRNA for ribosomal protein S20 in vitro. J. Mol. Biol. 234, 998–1012 (1993).
Stevens, A. An exoribonuclease from Saccharomyces cerevisiae: effect of modifications of 5′ end groups on the hydrolysis of substrates to 5′ mononucleotides. Biochem. Biophys. Res. Commun. 81, 656–661 (1978).
Acknowledgements
I thank G. Coburn for preparing degradosomes, A. Prud'homme-Genereux for helping to prepare Fig. 4, members of my laboratory for comments, and the MRC of Canada for an operating grant.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mackie, G. Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395, 720–724 (1998). https://doi.org/10.1038/27246
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/27246
This article is cited by
-
Relaxed Cleavage Specificity of Hyperactive Variants of Escherichia coli RNase E on RNA I
Journal of Microbiology (2023)
-
Circular RNAs and their role in renal cell carcinoma: a current perspective
Cancer Cell International (2021)
-
Biogenesis and functions of circular RNAs and their role in diseases of the female reproductive system
Reproductive Biology and Endocrinology (2020)
-
Structural and mechanistic basis of mammalian Nudt12 RNA deNADding
Nature Chemical Biology (2019)
-
A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.