Abstract
Cohesion between sister chromatids, mediated by the chromosomal cohesin complex, is a prerequisite for their alignment on the spindle apparatus and segregation in mitosis. Budding yeast cohesin first associates with chromosomes in G1. Then, during DNA replication in S-phase, the replication fork-associated acetyltransferase Eco1 acetylates the cohesin subunit Smc3 to make cohesin’s DNA binding resistant to destabilization by the Wapl protein. Whether stabilization of cohesin molecules that happen to link sister chromatids is sufficient to build sister chromatid cohesion, or whether additional reactions are required to establish these links, is not known. In addition to Eco1, several other factors contribute to cohesion establishment, including Ctf4, Ctf18, Tof1, Csm3, Chl1 and Mrc1, but little is known about their roles. Here, we show that each of these factors facilitates cohesin acetylation. Moreover, the absence of Ctf4 and Chl1, but not of the other factors, causes a synthetic growth defect in cells lacking Eco1. Distinct from acetylation defects, sister chromatid cohesion in ctf4Δ and chl1Δ cells is not improved by removing Wapl. Unlike previously thought, we do not find evidence for a role of Ctf4 and Chl1 in Okazaki fragment processing, or of Okazaki fragment processing in sister chromatid cohesion. Thus, Ctf4 and Chl1 delineate an additional acetylation-independent pathway that might hold important clues as to the mechanism of sister chromatid cohesion establishment.
Similar content being viewed by others
Introduction
Faithful segregation of chromosomes during mitosis requires sister chromatid cohesion from the time of DNA replication until mitosis. Cohesion is mediated by the essential chromosomal cohesin complex, a large ring-shaped protein complex composed of at least four subunits, Smc1, Smc3, Scc1 and Scc3 (Strunnikov et al. 1993; Guacci et al. 1997; Michaelis et al. 1997; Tóth et al. 1999). In budding yeast, cohesin is loaded onto chromosomes during late G1 by a loading complex consisting of the two Scc2 and Scc4 subunits (Ciosk et al. 2000; Lengronne et al. 2004). However, the association of cohesin with chromatin is not sufficient for sister chromatid cohesion. The formation of cohesive linkages between cohesin and the sister chromatids occurs during DNA replication in S-phase and depends on a number of ‘cohesion establishment factors’ (reviewed by Uhlmann 2009). Among these, the acetyltransferase Eco1 plays an essential role (Skibbens et al. 1999; Tóth et al. 1999; Ivanov et al. 2002). Eco1 is recruited to the replication fork probably by its physical interaction with the polymerase processivity factor PCNA (Lengronne et al. 2006; Moldovan et al. 2006), and acetylates cohesin’s Smc3 subunit during the progression of DNA replication (Ben-Shahar et al. 2008; Ünal et al. 2008; Zhang et al. 2008). As the consequence of Smc3 acetylation, cohesin’s DNA binding becomes resistant to the destabilizing effect of the cohesin-associated protein Wapl. In vertebrates, Smc3 acetylation promotes recruitment of sororin to the cohesin complex, which is thought to antagonize Wapl (Nishiyama et al. 2010). Whether a sororin paralog exists in yeast and how Smc3 acetylation counteracts Wapl is still not completely understood. In any event, the effect of Smc3 acetylation is an increased residence half-life of cohesin on chromosomes following DNA replication (Gerlich et al. 2006; Bernard et al. 2008; Chan et al. 2012; Lopez-Serra et al. 2013).
Stabilization of cohesin on chromosomes is likely a prerequisite for durable sister chromatid cohesion. On the other hand, whether cohesin stabilization is a sufficient explanation for establishment of sister chromatid cohesion is not known. If the replication fork is able to traverse through the large diameter of the cohesin ring, acetylating cohesin complexes that trap the newly synthesized sister chromatids along the way would be an efficient way to produce stable cohesion. Alternatively, cohesin might establish links between any pairs of DNA strands that come into its vicinity. In this case, cohesin acetylation close to the replication fork will again have a high probability to stabilize links between newly replicated DNA strands. Whether additional reactions are required to establish cohesin’s interaction with newly replicated sister chromatids in the first place is as yet unknown.
In addition to Eco1, several non-essential cohesion establishment factors contribute to sister-chromatid cohesion by as yet unknown mechanisms, including Ctf4, Ctf18, Tof1, Csm3, Chl1 and Mrc1 (Hanna et al. 2001; Mayer et al. 2001, 2004; Petronczki et al. 2004; Skibbens 2004; Warren et al. 2004). While these factors are non-essential for viability, their absence leads to compromised sister chromatid cohesion. Several lines of evidence have linked these cohesion establishment factors to the DNA replication fork, though their specific requirement for cohesion establishment during S-phase, as opposed to cohesin loading onto chromosomes or maintaining sister chromatid cohesion, has formally been demonstrated only for the Ctf18 complex and Csm3 (Xu et al. 2007). Both Ctf4 and Ctf18 have been detected at DNA replication forks (Lengronne et al. 2006), where Ctf4 is an integral component of the replisome progression complex (Gambus et al. 2006). Ctf4 binds to DNA polymerase α/primase (Miles and Formosa 1992; Zhou and Wang 2004) and mediates its interaction with the GINS complex (Gambus et al. 2009; Tanaka et al. 2009). Ctf18 is part of a replication factor C (RFCCtf18) complex that can both load and unload PCNA from DNA in vitro (Bermudez et al. 2003; Bylund and Burgers 2005), although its in vivo role, at least at hydroxyurea (HU)-stalled replication forks in budding yeast, appears to be that of a PCNA loader (Lengronne et al. 2006). Tof1, Csm3 and Mrc1 are also components of the replisome and share roles as regulators of replication fork pausing and activation of the DNA replication checkpoint, in addition to their role in sister chromatid cohesion (Alcasabas et al. 2001; Katou et al. 2003; Warren et al. 2004; Bando et al. 2009). Their molecular mechanism of action is still poorly understood. Chl1 was one of the first identified budding yeast genes with a role in chromosome segregation. It encodes a DNA helicase, but little is known about its molecular function during DNA replication or in sister chromatid cohesion (Gerring et al. 1990; Hirota and Lahti 2000; Petronczki et al. 2004; Skibbens 2004). A putative link of Chl1 to lagging strand DNA synthesis has been derived from its ability to bind PCNA and stimulate the catalytic activity of the Okazaki fragment processing flap endonuclease Fen1 in vitro (Farina et al. 2008). Ctf4 has also been implicated in lagging strand DNA synthesis as it recruits the DNA polymerase α/primase to the replication fork and due to its genetic interaction with the Dna2 helicase/nuclease, involved in Okazaki fragment processing (Formosa and Nittis 1999; Errico et al. 2009).
The above non-essential cohesion establishment factors have been grouped into two pathways, based on their genetic interactions in budding yeast (Xu et al. 2007). One group contains Ctf4, Tof1, Csm3 and Chl1, and the second the RFCCtf18 complex and Mrc1. However, the relationship of these two pathways to Eco1 has not yet been determined. Either or both of the pathways might act to promote Smc3 acetylation during replication fork progression or they might independently contribute to sister chromatid cohesion. In this study, we have used genetic and molecular assays to investigate the relationship of cohesion establishment factors and the cohesin acetylation pathway. This revealed that each of the cohesion establishment factors makes a contribution to cohesin acetylation. Removal of the cohesin destabilizer Wapl corrects the cohesion defect in most cohesion establishment mutants, but not in cells lacking Ctf4 or Chl1. The absence of Ctf4 or Chl1, unlike the others, causes pronounced synthetic growth defects in cells lacking Eco1 and Wapl. These findings suggest that, in addition to cohesin acetylation, Ctf4 and Chl1 act in an as yet uncharacterized, Eco1-independent cohesion establishment reaction. While Eco1 adds a lasting acetyl mark that stabilizes newly built sister chromatid cohesion, Ctf4 and Chl1 could hold a clue as to how links between the sister chromatids are established when the replisome meets cohesin during replication fork progression.
Materials and methods
Yeast strains and culture
All yeast strains used in this study were of W303 background. They are listed in Table S1. Epitope tagging and gene deletions were performed by gene targeting using polymerase chain reaction products (Wach et al. 1994; Knop et al. 1999). Cells were grown in YEP medium supplemented with either 2 % glucose or 2 % raffinose as the carbon source. To induce gene expression from the GAL1 promoter, 2 % galactose was added to cells grown in raffinose-containing medium. Cell synchronization in G1 was performed by addition of α-factor (0.4 μg/ml) or a-factor (0.04 μg/ml) for 2 h, as described (Lengronne et al. 2004; O'Reilly et al. 2012). To arrest cells in early S-phase, G1 synchronized cultures were filtered, washed and resuspended in fresh medium containing 0.1 M HU for 60 min. Mitotic arrest was achieved by release from G1 into medium containing 5 μg/ml nocodazole for 120 min. The use of the auxin-inducible degron (aid) was based on a previously published method (Nishimura et al. 2009).
Experimental techniques
The status of sister chromatid cohesion was analyzed by visualizing tetracycline repressor-GFP fusion proteins bound to tetracycline operator arrays integrated at the URA3 locus on chromosome 5, as previously described (Michaelis et al. 1997). For each condition, at least 100 cells were scored and each experiment was repeated three times. Means of the three biological replicates and the standard deviation are reported. Protein extracts were prepared from TCA fixed cells, separated by SDS-PAGE and analyzed by immunoblotting using a monoclonal α-acetyl-Smc3 antibody (a kind gift from K. Shirahige), α-HA clone 12CA5, α-Pk clone SV5-Pk1 (Serotec), a monoclonal α-Ctf4 antibody, and α-tubulin and α-Hmo1 sera (Abcam). Quantification of the Western blot signals was performed using a peroxidase-coupled secondary antibody and enhanced chemoluminescence measurement using a ImageQuant LAS 4000 biomolecular imager (GE Healthcare). Coimmunoprecipitation assays were performed from cell extracts prepared as described (Ben-Shahar et al. 2008), but SDS was omitted from the extraction buffer. Cell separation into soluble and chromatin-bound fractions was achieved from whole-cell extracts prepared by spheroplast lysis as previously described (Uhlmann et al. 1999). The assay to measure cohesin stability on chromosomes using the anchor-away technique followed a published procedure (Lopez-Serra et al. 2013), as did the chromatin immunoprecipitation assay (Lengronne et al. 2006) and the Okazaki fragment length analysis (Smith and Whitehouse 2012).
Results
Cohesion establishment factors promote cohesin acetylation
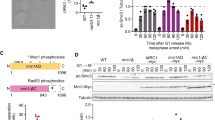
The relationship of the nonessential cohesion establishment factors, Ctf4, Ctf18, Tof1, Csm3, Chl1 and Mrc1, to Eco1-dependent cohesin acetylation is not yet known. They could aid Eco1 function, or they could contribute to sister chromatid cohesion independently of cohesin acetylation. The key target of Eco1 in cohesion establishment during S-phase in budding yeast is the cohesin subunit Smc3 (Ben-Shahar et al. 2008; Ünal et al. 2008), so we investigated whether any of the cohesion establishment factors contributed to Smc3 acetylation. We synchronized wild type cells and cells lacking each of the cohesion establishment factors (ctf4Δ, ctf18Δ, tof1Δ, csm3Δ, chl1Δ or mrc1Δ) in G1 using α-factor and released them to pass through S-phase and into a nocodazole-imposed mitotic arrest. In wild type cells, Smc3 acetylation accumulated during DNA replication in S-phase and persisted into mitosis, as seen before (Ben-Shahar et al. 2008; Ünal et al. 2008) (Fig. 1a). A similar pattern of Smc3 acetylation was detected in each of the cohesion establishment factor mutant strains, but the intensity of the acetyl-Smc3 signals appeared weaker in each case.
Cohesion establishment factors contribute to Smc3 acetylation. a Cells of the indicated genotypes were synchronized in G1 by α-factor treatment and released into nocodazole-imposed mitotic arrest. FACS analysis of the DNA content was used to monitor cell cycle progression. The Smc3 acetylation status was analyzed by Western blotting using an α-acetyl-Smc3 antibody. Total Smc3 levels served as the loading control and were detected using an antibody against its C-terminally fused Pk epitope. b The acetyl-Smc3 signal and loading control were quantified in three independent experiments and the mean and standard deviation of the normalized Smc3 acetylation levels are depicted. c Eco1 levels are comparable between wild type and cohesion establishment factor deficient cells. Cultures were synchronized in G1 using α-factor and released in hydroxyurea (HU)-containing medium. Eco1 protein levels were compared between the indicated strains by Western blotting against its HA epitope tag. Swi6 served as the loading control
To quantify the Smc3 acetylation level in each strain, we performed quantitative Western blotting of acetyl-Smc3 in three independent experiments, normalized to the total Smc3 detected via its Pk epitope tag (Fig. 1b). This revealed a reproducible reduction of Smc3 acetylation in most cohesion establishment factor mutant strains to between 45 % and 60 % of the levels observed in the wild type control. Cells lacking Ctf4 and Chl1 showed an even greater reduction, displaying only approximately 30 % of the wild type acetyl-Smc3 signal. This demonstrates that the cohesion establishment factors studied in this experiment contribute to cohesin acetylation during DNA replication, offering a possible explanation for defective sister chromatid cohesion in their absence.
A reason for decreased Smc3 acetylation could be reduced Eco1 levels. We therefore compared Eco1 levels between wild type and cohesion establishment factor mutant strains, after arrest in early S-phase using HU. This revealed comparable amounts of Eco1 in all strains (Fig. 1c). Eco1 levels appeared slightly lower in ctf4Δ and ctf18Δ cells, but this is likely due to the difficulty of synchronizing these cells by HU treatment. Eco1 becomes unstable once cells progress out of S-phase into G2 (Borges et al. 2010; Lyons and Morgan 2011). We conclude that lower Smc3 acetylation in cohesion factor establishment mutant strains is unlikely explained by reduced Eco1 levels. The cohesion establishment factors appear to facilitate Smc3 acetylation by Eco1 via a mechanism that merits further investigation.
Cohesin acetylation levels do not strictly correlate with sister chromatid cohesion
The sister chromatid cohesion defect seen in cells lacking Ctf4 or Ctf18 is of comparable severity (Hanna et al. 2001; Xu et al. 2007; and see below). Yet, acetyl-Smc3 levels were lower in ctf4Δ cells compared to ctf18Δ cells. We therefore investigated the extent to which reduced Smc3 acetylation correlates with an accompanying sister chromatid cohesion defect by including the temperature sensitive eco1-1 mutant in our analysis. Smc3 acetylation is much reduced in cells carrying this allele, even at the permissive temperature, and becomes close to undetectable at the restrictive temperature (Rowland et al. 2009). We synchronized cells in G1 and followed them through S-phase and into arrest in mitosis by nocodazole treatment at 23 °C, a permissive temperature for the eco1-1 allele (Fig. 2a). At the time of S-phase, Smc3 acetylation became detectable in eco1-1 cells, though at a greatly reduced level compared to wild type cells. Acetylation remained markedly lower even than in ctf4Δ and chl1Δ cells. When analyzing sister chromatid cohesion, we found that it was compromised in the ctf4Δ and chl1Δ strains to a similar extent when compared to the eco1-1 strain at permissive temperature (Fig. 2b). These findings suggest that reduced Smc3 acetylation in the ctf4Δ and chl1Δ cells likely contributes to, but might not fully explain, the severity of the observed cohesion defect.
Cohesin acetylation levels and cohesion defect do not strictly correlate. a Smc3-Pk was immunopurified from extracts of the indicated cells, progressing through a synchronous cell cycle following α-factor block and release into nocodazole-imposed mitotic arrest at 23 °C. The Smc3 acetylation status was analyzed by Western blotting. FACS analysis of the DNA content was used to monitor cell cycle progression. b Cells of the indicated genotypes were synchronized in G1 using α-factor and released into nocodazole-imposed mitotic arrest at 23 °C. Sister chromatid cohesion at the GFP-marked URA3 locus was analyzed
Ctf4 and Chl1 define a subset of Eco1-independent cohesion establishment factors
Given the limitation of Smc3 acetylation levels as a readout for cohesion defects, we turned towards a genetic analysis of the relationship between cohesion establishment factors and the Eco1-dependent cohesion establishment reaction. Factors that act by promoting cohesin acetylation should no longer affect cell fitness in cells lacking Eco1, for example in the eco1Δ wpl1Δ strain background in which cohesin acetylation is dispensable for cell viability. If, on the other hand, a cohesion establishment factor acts in a different pathway from Eco1, we would expect to see a synthetic growth defect when deleting it from eco1Δ wpl1Δ cells. We were able to obtain ctf18Δ, tof1Δ csm3Δ, chl1Δ and mrc1Δ deletions in an eco1Δ wpl1Δ strain background and compared the growth of the resulting triple mutant strains to that of the parental strains (Fig. 3a). After streaking on rich medium plates and incubation at 25 °C or 30 °C, the growth of most triple mutant strains was not greatly different from that of the eco1Δ wpl1Δ double mutant. An exception was observed in the case of chl1Δ. While the chl1Δ strain by itself grew robustly, the chl1Δ deletion caused poor growth in the eco1Δ wpl1Δ background at 25 °C, which was even more pronounced at 30 °C. Therefore, chl1Δ, but not the other establishment factor deletions, caused a marked synthetic growth defect in the absence of eco1. This suggests that Chl1 acts, at least in part, in a pathway parallel to Smc3 acetylation.
Ctf4 and Chl1 define a subset of Eco1-independent cohesion establishment factors. a chl1Δ deletion, but not other cohesion establishment factor deletions, causes a synthetic growth defect in the eco1Δ wpl1Δ background. Strains of the indicated genotypes were streaked on YPD medium and incubated at the indicated temperatures for 2–3 days. b Ctf4 is essential in the eco1Δ wpl1Δ background. A ctf4Δ/CTF4 eco1Δ/ECO1 wpl1Δ/WPL1 heterozygous diploid was sporulated, and the genotype of the viable spores in each tetrad was determined. Inferred genotypes of unviable spores are in gray. Asterisks (*) denote pairs of spores with either of the two indicated genotypes. c Search for additive cohesion defects in a eco1Δ wpl1Δ strain. Strains of the indicated genotypes were synchronized in G1 by α-factor block and released into nocodazole-imposed mitotic arrest. Sister chromatid cohesion at the GFP-marked URA3 locus was analyzed in 100 cells. The mean and standard deviation from three experiments are indicated. A dashed line marks the cohesion defect of the eco1Δ wpl1Δ strain. A binomial test showed that the cohesion defect differences of strains containing additional establishment factor deletions were not statistically significant (ns). d Rescue of the cohesion defect in most cohesion establishment factor mutants, but not ctf4Δ and chl1Δ, by wpl1Δ deletion. Strains of the indicated genotypes were synchronized in G1 by α-factor block and released into nocodazole-imposed mitotic arrest. Sister chromatid cohesion was analyzed as in c. The significance of the sister chromatid cohesion rescue by wpl1Δ deletion was analyzed using a binomial test (*p < 0.05; **p < 0.005; ns, not significant)
We were unable to delete CTF4 in the eco1Δ wpl1Δ strain background by gene targeting. To analyze whether this was because a ctf4Δ eco1Δ wpl1Δ triple mutant strain is unviable, we analyzed tetrads after dissection of a ctf4Δ/CTF4 eco1Δ/ECO1 wpl1Δ/WPL1 heterozygous diploid (Fig. 3b). Genotyping of the viable spores revealed that the ctf4Δ eco1Δwpl1Δ combination was not viable while eco1Δ wpl1Δ and also ctf4Δ wpl1Δ strains were recovered at the expected frequency. In an additional attempt to obtain a ctf4Δ eco1Δ wpl1Δ strain, we introduced the CTF4 gene on an episomal plasmid that also contained the URA3 selectable marker. After deleting the genomic CTF4 locus we tested whether the plasmid-borne CTF4 gene could be lost by counterselection against the URA3 gene on medium containing 5-fluoroorotic acid (5-FOA). Unlike from a ctf4Δ control strain, the CTF4-containing plasmid could not be lost from the ctf4Δ eco1Δ wpl1Δ strain and no 5-FOA-resistant colonies were recovered (Fig. S1). This confirms that Ctf4 becomes essential in the absence of Eco1. It suggests that, like Chl1, Ctf4 plays a role that is at least in part independent of promoting Eco1-dependent cohesin acetylation.
We next wanted to address whether the synthetic growth defect of ctf4Δ and chl1Δ in the eco1Δ wpl1Δ background was due to an additive effect on sister chromatid cohesion, or due to an unrelated reason. We therefore compared cohesion in metaphase arrested eco1Δ wpl1Δ cells to the viable triple mutants lacking the additional cohesion establishment factors. eco1Δ wpl1Δ cells by themselves display a considerable cohesion defect (Ben-Shahar et al. 2008). After release from G1 into nocodazole-imposed mitotic arrest, approximately 35 % of cells show split GFP-marked URA3 loci. Cells containing the single cohesion establishment factor deletions ctf18Δ, tof1Δ, csm3Δ, chl1Δ and mrc1Δ showed between 20 to 30 % premature sister chromatid splitting. However, their additional deletion did not significantly alter the cohesion defect in the eco1Δ wpl1Δ background (Fig. 3c). This included the chl1Δeco1Δwpl1Δ triple mutant strain that had shown a marked synthetic growth defect. As a positive control for an increased cohesion defect, we reintroduced Wapl into the eco1Δ wpl1Δ strain by expressing it from the inducible GAL1 promoter at the time of G1 release. This should expose the full cohesion defect of an eco1Δ strain but caused only a small increase in the percentage of split GFP signals. This suggests that under our experimental conditions an additional cohesion defect in the eco1Δ wpl1Δ strain background, either by reintroducing Wapl or by additional cohesion establishment factor deletions, is difficult to ascertain.
We therefore took an alternative approach to test whether the Eco1-independent function of Ctf4 and Chl1 lies in sister chromatid cohesion. Cohesion defects due to defective Smc3 acetylation are partly rescued by the absence of Wapl. This has been documented in both budding yeast and human cells (Gandhi et al. 2006; Ben-Shahar et al. 2008; Chan et al. 2012; Lopez-Serra et al. 2013). In contrast, if Chl1 and Ctf4 act in sister chromatid cohesion independently of cohesin acetylation, then the cohesion defect in their absence should not be rescued in this manner. To investigate this, we introduced the wpl1Δ deletion into strains lacking each cohesion establishment factor and again measured sister chromatid cohesion after cell synchronization. The cohesion defects in ctf18Δ, tof1Δ, csm3Δ and mrc1Δ strains were significantly rescued by the wpl1Δ deletion, to levels close to that of the wpl1Δ strain itself (Fig. 3d). In contrast, the cohesion defects in ctf4Δ and chl1Δ cells remained unaffected in the absence of Wapl. These results are consistent with the idea that Ctf18, Tof1, Csm3 and Mrc1 act to support Eco1-dependent cohesin stabilization, while Chl1 and Ctf4 in addition act in an Eco1-independent pathway to establish sister chromatid cohesion.
Ctf4, Chl1 and cohesin stabilization on chromosomes
The consequence of Eco1-dependent Smc3 acetylation is stabilization of cohesin’s binding to chromosomes (Bernard et al. 2008; Chan et al. 2012; Lopez-Serra et al. 2013). If Ctf4 and Chl1 act in an Eco1-independent cohesion establishment pathway, their absence should have little impact on the stability of cohesin’s DNA binding. To investigate this, we measured cohesin’s residence time on chromosomes using an assay based on the ‘anchor-away’ technique, as previously described (Haruki et al. 2008; Lopez-Serra et al. 2013). In brief, we fused the cohesin subunit Scc1 in tandem to GFP, for visualization, and to FRB, half of a pair of rapamycin-inducible protein interaction domains. The other half, FKBP12, was attached to the ribosomal protein Rpl13a. By hitchhiking onto Rpl13a while it shuttles through the nucleus during ribosome assembly, a freely diffusible nuclear protein is quickly depleted from the nucleus following rapamycin addition (Haruki et al. 2008; Fig. 4a). This is the case for cohesin before DNA replication when it turns over on chromosomes due to the activity of Wapl, but not afterwards when it reaches stable binding due to acetylation by Eco1 (Lopez-Serra et al. 2013).
Effect of cohesion establishment mutants on cohesin stability on chromosomes. a Schematic of the anchor-away experiment to measure the persistence of nuclear Scc1-GFP enrichment as readout for cohesin stability on chromosomes. b Strains of the indicated genotypes were synchronized in G1 by α-factor block. 500 μM auxin was added to the growth medium to degrade Eco1-aid before release into nocodazole-imposed mitotic arrest. Next, 1 μM rapamycin was added (+r) and samples taken in 10-min intervals (compare Lopez-Serra et al. 2013). FACS analysis of DNA content is shown to monitor cell cycle progression. The fraction of cells with visible nuclear Scc1-GFP retention is indicated
Since the host strain for the anchor-away technique is of the α mating type, we synchronized cells in G1 using a-factor block and release before arresting them in metaphase by nocodazole treatment (O'Reilly et al. 2012). We now added rapamycin and assessed the nuclear enrichment of Scc1-FRB-GFP in 10-min intervals. In wild type cells, Scc1 is stably bound to chromosomes and therefore retains nuclear localization throughout the experiment (Fig. 4b). As reported, after Eco1 depletion using an auxin-inducible degron, Scc1 was completely lost from the nucleus within 60 min of rapamycin addition (Lopez-Serra et al. 2013). Absence of either Ctf4 or Chl1 led only to a slight decrease in the stability of nuclear Scc1, when compared to the wild type, consistent with a role independent of promoting cohesin stability. However, ctf4 and chl1 deletions cause a less severe cohesion defect compared to eco1 inactivation. We therefore analyzed cells lacking Ctf18, a component of the Eco1-dependent pathway, that show a cohesion defect comparable to that of cells lacking Ctf4 or Chl1. Cohesin in ctf18Δ cells was also only partly destabilized, although reproducibly to a greater extent than in ctf4Δ and chl1Δ cells. These results are consistent with the possibility that the cohesion defect seen in the absence of Ctf4 and Chl1 is in part independent of the role of these proteins in promoting Smc3 acetylation.
Molecular characterization of Ctf4 and Chl1 in sister chromatid cohesion
Ctf4 and Chl1 have previously been recognized to be genetically related cohesion establishment factors (Xu et al. 2007). While Ctf4 act as a hub of protein interactions within the replisome progression complex (Miles and Formosa 1992; Zhou and Wang 2004; Gambus et al. 2006, 2009; Tanaka et al. 2009), little is known about the molecular function of Chl1. We first confirmed the close genetic relationship between Ctf4 and Chl1 by comparing the cohesion defects in the single ctf4Δ and chl1Δ and double ctf4Δ chl1Δ mutant cells (Fig. 5a). As reported (Xu et al. 2007), we could not observe an increase of the cohesion defect after combining the two single mutants, suggesting that the two proteins closely collaborate in the same pathway of cohesion establishment.
Relationship between Ctf4, Chl1 and cohesin. a No additive cohesion defects when combining ctf4Δ and chl1Δ deletions. Strains of the indicated genotypes were synchronized in G1 by α-factor block and released into nocodazole-imposed mitotic arrest. Sister chromatid cohesion at the GFP-marked URA3 locus was analyzed. b Ctf4 and Chl1 are not part of a stable protein complex. Epitope-tagged Chl1 or Pol1 were immunoprecipitated using an α-Pk antibody and coprecipitation of Ctf4 was analyzed by immunoblotting. Whole cell extracts (WCE) and immunoprecipitates (IP) are shown. c chl1Δ deletion reduces cohesin association with chromosomes. Wild type and chl1Δ cells were arrested in mitosis by nocodazole treatment. Cells were processed for chromatin immunoprecipitation against the Pk epitope-tagged cohesin subunit Scc1. Chromatin immunoprecipitates were analyzed by quantitative PCR at three cohesin binding sites at convergent intergenic regions on chromosome arms and three centromeres. Mean and standard deviation of three repeats of the experiment are shown. d Cells of the indicated genotypes were synchronized in G1 by α-factor treatment and released into either HU or nocodazole (NOC)-containing media. Aliquots of the cultures were taken before synchronization (cycling cells [cyc]). Whole cell extracts (WE) were separated into supernatant (SU) and chromatin (CP) fractions, and Scc1-Pk was detected by immunoblotting. Tubulin and Hmo1 served as loading controls for the supernatant and chromatin fractions, respectively
A common mechanism by which two proteins act in the same pathway is that they are part of a protein complex. We therefore asked whether we could find evidence for a direct protein interaction between Ctf4 and Chl1. To do this, we analyzed coimmunoprecipitation of Ctf4 and Chl1, fused at their C termini to an HA and a Pk epitope, respectively. However, immunoprecipitation of Chl1 via its Pk epitope did not coprecipitate any detectable Ctf4 (Fig. 5b). As a control, we fused the DNA polymerase α/primase subunit Pol1 to a Pk epitope. Its pulldown efficiently coprecipitated Ctf4, consistent with previous reports (Miles and Formosa 1992; Zhou and Wang 2004). Therefore, Ctf4 and Chl1 do not engage in a stable physical interaction, at least not one that is detectable by our coimmunoprecipitation assay.
To gain insight into the mechanism of Ctf4 and Chl1 action, we investigated whether they are recruited to chromatin in a mutually dependent manner. For this, we separated whole cell extracts into soluble and chromatin-bound fractions at different stages of the cell cycle and analyzed the distribution of Ctf4 and Chl1 by Western blotting (Fig. S2). Ctf4 binding to chromatin was weak in G1, but increased in S-phase arrested cells, consistent with its function as part of the replisome progression complex (Gambus et al. 2006). Ctf4 chromatin binding in S-phase depended on Chl1. In contrast, Chl1 binding to chromatin appeared constant throughout the different stages of the cell cycle and was independent of Ctf4. These findings suggest that Chl1 is a constitutive chromosomal component and open the possibility that Chl1 might act to facilitate Ctf4 recruitment. A limitation of the chromatin fractionation technique is that any Triton X-100-insoluble structure is seen as part of the chromatin pellet. Future experiments using chromatin immunoprecipitation analyses will be required to understand when and where Chl1 associates with chromosomes.
Chl1 promotes cohesin binding to DNA
A recent report suggested that Chl1 promotes cohesin loading at the centromere of chromosome 3 (Laha et al. 2011). To confirm these observations and to test whether Chl1 also promotes cohesin binding to other chromosomal loci, we performed ChIP analysis of the Pk epitope-tagged cohesin subunit Scc1 in a wild type and chl1Δ strain background. Quantitative analysis of the chromatin immunoprecipitate by real time PCR revealed an up to 2-fold reduced cohesin level at four out of six centromere and chromosome arm loci tested (Fig. 5c). A reduced ChIP signal could be due to decreased cohesin loading, or due to overall reduced cohesin levels in chl1Δ cells. To differentiate between these possibilities, we analyzed Scc1 levels in whole cell extracts, supernatant and chromatin fractions prepared from wild type and chl1Δ cells (Fig. 5d). This showed that total Scc1 levels in the whole cell extract were comparable between chl1Δ and wild type cells, but that chromatin binding of Scc1 was noticeably reduced in the absence of Chl1. Decreased cohesin levels on chromosomes are in itself unlikely sufficient to explain the cohesion defect in cells lacking Chl1. This is because it has been shown that even a greater than 7-fold reduction in cohesin levels does not cause a cohesion defect, if Chl1 is active (Heidinger-Pauli et al. 2010). Nevertheless, altered chromosomal cohesin levels could be a hint as to the function of Chl1 in cohesion establishment. In the future, it will be important to define at which cell cycle stage Chl1 affects cohesin loading, and whether this function is shared with Ctf4.
Okazaki fragment processing and sister chromatid cohesion
Establishment of sister-chromatid cohesion is tightly coupled to DNA replication during S-phase (Uhlmann and Nasmyth 1998; Lengronne et al. 2006). Both Ctf4 and Chl1 have previously been implicated in lagging strand DNA synthesis and Okazaki fragment maturation (Miles and Formosa 1992; Formosa and Nittis 1999; Zhou and Wang 2004; Farina et al. 2008; Gambus et al. 2009; Tanaka et al. 2009; Rudra and Skibbens 2012). If the ‘trombone model’ for Okazaki fragment synthesis is applicable to eukaryotic DNA replication, the lagging strand loop could be a potential obstacle if replication forks were to pass through cohesin rings (Hamdan and Richardson 2009; Uhlmann 2009). In this model, Ctf4 and Chl1 could act to coordinate fork passage through cohesin rings with Okazaki fragment processing, e.g., to ensure lagging strand loop release upon cohesin encounter, or to ensure that loops do not exceed a maximum size. We therefore investigated the interplay between Ctf4 and Chl1, Okazaki fragment processing and sister chromatid cohesion.
It has been reported that cells lacking Ctf4 show synthetic lethality with the dna2-2 mutation (Formosa and Nittis 1999; and data not shown). Dna2 is a nuclease-helicase involved in Okazaki fragment processing, whose function overlaps with that of the flap endonuclease Fen1 (Budd and Campbell 1997). Human Fen1, in turn, has been shown to be required for sister chromatid cohesion (Farina et al. 2008). If the synthetic growth defect of ctf4Δ with dna2-2 is due to Dna2’s role in Okazaki fragment processing, we would expect a similar synthetic interaction between ctf4Δ and fen1Δ mutations. We therefore analyzed the haploid progeny after sporulation of a heterozygous diploid ctf4Δ/CTF4 fen1Δ/FEN1 strain. This revealed that a ctf4Δ fen1Δ double mutant strain is not viable (Fig. 6a). Likewise, we were unable to obtain a viable chl1Δ fen1Δ strain. We were able to obtain viable, but poorly growing, chl1Δ dna2-2 spores. These genetic interactions are consistent with a link between Okazaki fragment processing and sister chromatid cohesion, or they could alternatively arise due to an overlapping role of these proteins in another process relating to DNA replication or repair.
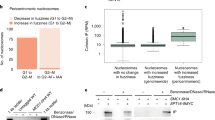
Relationship of Ctf4, Chl1 and Okazaki fragment processing. a Synthetic lethality of chl1Δ and ctf4Δ with fen1Δ and poor growth of chl1Δ in combination with the dna2-2 allele. Heterozygous diploid strains of the indicated genotypes were sporulated and the genotype of the viable spores in each tetrad was determined. Inferred genotypes of unviable spores are in gray. b Budding yeast cells lacking Fen1, or carrying the dna2-2 allele, show intact sister chromatid cohesion. Strains of the indicated genotypes were synchronized in G1 by α-factor block and released into nocodazole-imposed mitotic arrest. Sister chromatid cohesion at the GFP-marked URA3 locus was analyzed. fen1Δ and ctf18Δ cells, for comparison, were grown at 25 °C, dna2-2 cells were released from G1 at 37 °C. A wild type control at each temperature is included. c Unchanged Okazaki fragment length distribution in cells lacking Ctf4, Chl1 or Ctf18. DNA ligase I was inactivated in the indicated strain backgrounds and the Okazaki fragment length distribution analyzed as described (Smith and Whitehouse 2012). An intensity scan of each lane is included and the position of mono-, di-, and trinucleosome sized fragments is indicated. For comparison, cac1Δ cells were analyzed, in which Okazaki fragments are longer. d Increased Okazaki fragment length, observed in cells with compromised replication-coupled chromatin assembly, does not cause a sister chromatid cohesion defect. Strains of the indicated genotypes were synchronized in G1 by α-factor block and released into nocodazole-imposed mitotic arrest at 25 °C. Sister chromatid cohesion at the GFP-marked URA3 locus was analyzed
To determine the contribution of Fen1 and Dna2 to sister chromatid cohesion, we analyzed the cohesion status in cells lacking Fen1 or carrying the dna2-2 mutation. Cells were synchronized in G1 by pheromone treatment and released to pass through a synchronous cell cycle before arrest in mitosis by nocodazole treatment. Sister chromatid cohesion at the URA3 locus was scored and wild type and ctf18Δ cells were included as controls (Fig. 6b). Unlike in ctf18Δ cells, we could not observe any noticeable defect in sister chromatid cohesion in fen1Δ and dna2-2 cells. This is in marked contrast to human cells, in which Fen1 depletion by RNA interference causes a marked cohesion defect (Farina et al. 2008). In conclusion, we could not find evidence for a conserved role of the Okazaki fragment processing enzymes Fen1 and Dna2 in sister chromatid cohesion.
We next examined whether the interaction of Ctf4 with DNA polymerase α/primase indicates a role in regulating Okazaki fragment priming frequency and consequently Okazaki fragment length. We utilized a recently developed technique to visualize Okazaki fragments after inactivation of DNA ligase I (Smith and Whitehouse 2012). Because Ctf4 acts in the same genetic pathway as Chl1 we included both ctf4Δ and chl1Δ cells in the analysis, as well as ctf18Δ cells in which we would not expect Okazaki fragment length to be affected. In wild type cells, Okazaki fragments show a distinctive size distribution characterized by nucleosomal length units (Fig. 6c). This pattern and its size distribution was unaltered in ctf4Δ, chl1Δ and ctf18Δ cells. This is in contrast to cac1Δ cells, lacking CAF-1 chromatin assembly factor activity, in which Okazaki fragments are longer (Smith and Whitehouse 2012). Our assay measures the length distribution of mature Okazaki fragments, just before their ligation. We therefore cannot exclude Okazaki fragment priming differences in the absence of Ctf4. Nevertheless, the absence of a visible impact on the Okazaki fragment length distribution makes it unlikely that lagging strand loop size differs greatly in the presence or absence of Ctf4 and Chl1. We therefore consider it unlikely that the cohesion defect in ctf4Δ and chl1Δ cells is due to collisions of an excessively large lagging strand loop with cohesin rings.
Lastly, we addressed whether longer Okazaki fragment length would interfere with sister chromatid cohesion establishment. Termination of Okazaki fragment DNA synthesis is coupled to chromatin assembly and the average Okazaki fragment length substantially increases if the chromatin assembly factor CAF-1 is inactive (Smith and Whitehouse 2012; Fig. 6c). We therefore assessed sister chromatid cohesion in cells lacking the CAF-1 subunits Cac1 or Cac2, or lacking another factor implicated in replication-coupled chromatin assembly, Asf1. In neither cac1Δ, cac2Δ, nor asf1Δ cells could we detect a measureable sister chromatid cohesion defect (Fig. 6d). We currently do not know whether longer Okazaki fragments due to compromised replication-coupled chromatin assembly result in increased lagging strand loop size, or whether indeed Okazaki fragment synthesis in eukaryotes is accompanied by lagging strand loop formation. Nevertheless, we conclude that the only currently known situation in which Okazaki fragment length is increased did not interfere with the establishment of sister chromatid cohesion.
Discussion
The molecular mechanism underlying the establishment of sister chromatid cohesion during S-phase is still unknown, even though the activity of the only known essential contributor, Eco1, is now relatively well understood (Ben-Shahar et al. 2008; Bernard et al. 2008; Ünal et al. 2008; Zhang et al. 2008; Chan et al. 2012; Lopez-Serra et al. 2013). Eco1 acetylates cohesin’s Smc3 subunit to prevent the Wapl protein from destabilizing the interaction of the cohesin ring with DNA. But how does cohesin stabilization contribute to cohesion establishment, and what additional reactions might be required?
The contribution of Eco1 to cohesion establishment
Eco1-dependent acetylation of cohesin occurs during S-phase and depends on DNA replication fork progression at this time (Ben-Shahar et al. 2008). Several lines of circumstantial evidence place Eco1 at the replication fork. Its S-phase specific chromosome binding depends on PCNA, an auxiliary factor of DNA replication reactions that accumulates at replication forks (Moldovan et al. 2006). In addition, Eco1 has been placed at replication forks by chromatin immunoprecipitation analyses, at least to forks that were arrested in early S-phase by HU treatment (Lengronne et al. 2006). It still needs to be formally investigated whether Eco1 also localizes to replication forks in the process of undisturbed DNA replication, though that seems likely. It also remains to be explored exactly when and where, relative to the passing replication fork machinery, Eco1 places the Smc3 acetylation marks onto cohesin. Knowing the answers to these questions will help to understand the mechanism of replication-coupled Smc3 acetylation. As far as sister chromatid cohesion establishment is concerned, the exact timing of acetylation might not be crucial. Even non-acetylated cohesin rings likely sustain sister chromatid cohesion for a short while, giving a window of opportunity for cohesin acetylation. A previous suggestion that Smc3 acetylation is needed at the moment of DNA replication to facilitate replication fork progression in the face of a ‘cohesion anti-establishment’ activity that Wapl imposes (Rowland et al. 2009; Sutani et al. 2009; Terret et al. 2009) has not been confirmed in more recent studies (Chan et al. 2012; Lopez-Serra et al. 2013).
It seems plausible that stabilization of cohesin’s DNA binding is a prerequisite for sister chromatid cohesion. Without it, sister chromatid cohesion would deteriorate as cohesin turns over on chromosomes. This would be a problem especially at centromeres, where spindle forces start to pull sister chromatids away from each other and stable sister chromatid cohesion is required to resist these forces. Acetylation stabilizes cohesin by counteracting the destabilizing activity of Wapl. But even in the absence of Wapl, cohesin acetylation contributes to sister chromatid cohesion by stabilizing cohesin on chromosomes (Ben-Shahar et al. 2008; Lopez-Serra et al. 2013). This makes Smc3 acetylation a crucial cohesin modification during the creation of durable sister chromatid cohesion.
In the absence of other known essential factors, is cohesin acetylation sufficient to explain the establishment of sister chromatid cohesion? Cohesin is loaded onto DNA already before DNA replication. So how does the transition from binding one DNA strand to holding together two replicated strands occur? One possibility is that the replication fork simply passes through cohesin rings (Haering et al. 2002). In principle, the large diameter of cohesin would accommodate passage of all known replisome components (Lengronne et al. 2006), although we do not yet know how a eukaryotic replisome is assembled and whether the fully assembled replisome really fits through the cohesin ring. If it did, then indeed acetylation-dependent cohesin stabilization by replication fork-associated Eco1, as the fork passes through cohesin rings, could be the only essential cohesion establishment reaction.
An Eco1-independent cohesion establishment pathway
In addition to Eco1, numerous other cohesion establishment factors have been identified that contribute to sister chromatid cohesion. Many of them have been linked to the DNA replication fork, though it is important to point out that the execution time when these factors carry out their function has so far only been rigorously studied in the case of Ctf8, a component of the RFCCtf18 complex, and Csm3 (Xu et al. 2007). Both proteins become dispensable after S-phase, thus qualifying them as sister chromatid cohesion establishment factors that are no longer required for cohesion maintenance. We now provide evidence that many of these cohesion establishment factors, Ctf4, Ctf18, Tof1, Csm3, Chl1 and Mrc1 promote cohesin acetylation during S-phase. In the case of RFCCtf18, this could be due to its activity as a PCNA loader, which in turn probably serves as a recruitment platform for Eco1 at the replication fork. How the other factors contribute to Smc3 acetylation is less clear, they could also be involved in recruiting Eco1 to replication forks, or they could modulate Eco1’s activity at the fork. Once added, the Smc3 acetylation mark remains protected from deacetylation by the protein deacetylase Hos1 for as long as cohesin remains chromosome bound (Borges et al. 2010). The mechanism behind this protection is as yet unknown. It is therefore conceivable that cohesion establishment factors act at the level of either promoting acetylation or by establishing protection from deacetylation.
The two cohesion establishment factors Ctf4 and Chl1 are distinct from the others. Their deletion caused a strong synthetic growth defect in eco1Δ wpl1Δ cells. In addition, the cohesion defect in cells lacking Ctf4 or Chl1, unlike in cells lacking any of the other factors, was not ameliorated by removal of Wapl. This suggests that Ctf4 and Chl1 play a role in the establishment of sister chromatid cohesion that is independent of stabilizing chromosome binding of the cohesin complex. What could their role be? Ctf4 is an integral component of the replisome progression complex, engaged in protein interactions between the MCM DNA helicase, the GINS complex and the DNA polymerase α/primase complex. Chl1 in turn might contribute to Ctf4 recruitment during S-phase. If the replisome passes through cohesin rings during DNA replication, it is conceivable that an altered replisome geometry in the absence of Ctf4 could lead to difficulties with cohesion establishment. Instead of traversing through the ring, the oncoming replication fork might displace or break cohesin.
An alternative scenario for the establishment of sister chromatid cohesion is that the replisome does not pass through cohesin rings. In this case, cohesion might be established in a reaction that is similar to chromosome condensation by a relative of cohesin, the condensin complex. Condensin most likely acts by establishing DNA interactions between its binding sites on DNA strands that happen to come into proximity (D'Ambrosio et al. 2008; Haeusler et al. 2008; Thadani et al. 2012). Cohesin also takes part in chromosome condensation (Guacci et al. 1997; Lopez-Serra et al. 2013), which could be the legacy of the evolutionary relationship between the two complexes. In this scenario cohesin, like condensin, engages in a multitude of DNA interactions along and between sister chromatids. During DNA replication, when the two sister chromatids are juxtaposed as they emerge from the replisome, the two closest DNA strands are the two newly synthesized sister chromatids. Stabilization of cohesin at this time, by replication fork-associated Eco1, would have a good chance of stabilizing cohesin interactions between these sister chromatids. In this model of cohesion establishment, an important aspect would be the establishment of new DNA contacts by cohesin in the wake of the DNA replication fork. As yet, there is no evidence that the canonical cohesin loader Scc2/Scc4 is required at this time (Lengronne et al. 2006). However, we and others have now observed a defect in cohesin loading in the absence of Chl1 (Laha et al. 2011). Whether cells lacking Chl1 are specifically defective in establishing new contacts of cohesin with DNA during DNA replication, and whether Ctf4 shares this role, will be important questions to be addressed in the future. A better appreciation of where and when Ctf4 and Chl1 perform their role in sister chromatid cohesion establishment will be important to understand how cohesin establishes molecular links between sister chromatids.
References
Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJH, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ (2001) Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol 3:958–965
Bando M, Katou Y, Komata M, Tanaka H, Itoh T, Sutani T, Shirahige K (2009) Csm3, Tof1, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks. J Biol Chem 284:34355–34365
Ben-Shahar TR, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F (2008) Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321:563–566
Bermudez VP, Maniwa Y, Tappin I, Ozato K, Yokomori K, Hurwitz J (2003) The alternative Ctf18–Dcc1–Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc Natl Acad Sci USA 100:10237–10242
Bernard P, Schmidt CK, Vaur S, Dheur S, Drogat J, Genier S, Ekwall K, Uhlmann F, Javerzat JF (2008) Cell-cycle regulation of cohesin stability along fission yeast chromosomes. EMBO J 27:111–121
Borges V, Lehane C, Lopez-Serra L, Flynn H, Skehel M, Rolef Ben-Shahar T, Uhlmann F (2010) Hos1 deacetylates Smc3 to close the cohesin acetylation cycle. Mol Cell 39:677–688
Budd ME, Campbell JL (1997) A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol Cell Biol 17:2136–2142
Bylund GO, Burgers PMJ (2005) Replication protein A-directed unloading of PCNA by the Ctf18 cohesion establishment complex. Mol Cell Biol 25:5445–5455
Chan K-L, Roig MB, Hu B, Beckouët F, Metson J, Nasmyth K (2012) Cohesin's DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell 150:961–974
Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K (2000) Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5:1–20
D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F (2008) Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev 22:2215–2227
Errico A, Cosentino C, Rivera T, Losada A, Schwob E, Hunt T, Costanzo V (2009) Tipin/Tim1/And1 protein complex promotes Polα chromatin binding and sister chromatid cohesion. EMBO J 28:3681–3692
Farina A, Shin J-H, Kim D-H, Bermudez VP, Kelman Z, Seo Y-S, Hurwitz J (2008) Studies with the human cohesion establishment factor, ChlR1. J Biol Chem 283:20925–20936
Formosa T, Nittis T (1999) Dna2 mutants reveal interactions with Dna polymerase α and Ctf4, a Pol α accessory factor, and show that full Dna2 helicase activity is not essential for growth. Genetics 151:1459–1470
Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K (2006) GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8:358–366
Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, Calzada A, Labib K (2009) A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase α within the eukaryotic replisome. EMBO J 28:2992–3004
Gandhi G, Gillespie PJ, Hirano T (2006) Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol 16:2406–2417
Gerlich D, Koch B, Dupeux F, Peters J-M, Ellenberg J (2006) Live-cell imaging reveals a stable cohesin–chromatin interaction after but not before DNA replication. Curr Biol 16:1571–1578
Gerring SL, Spencer F, Hieter P (1990) The CHL1(CTF1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J 9:4347–4358
Guacci V, Koshland D, Strunnikov A (1997) A direct link between sister chromatid cohesion and chromosome condensation revealed through analysis of MCD1 in S. cerevisiae. Cell 91:47–57
Haering CH, Löwe J, Hochwagen A, Nasmyth K (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 9:773–788
Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR (2008) Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev 22:2204–2214
Hamdan SM, Richardson CC (2009) Motors, switches, and contacts in the replisome. Annu Rev Biochem 78:205–243
Hanna JS, Kroll ES, Lundblad V, Spencer FA (2001) Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol 21:3144–3158
Haruki H, Nishikawa J, Laemmli UK (2008) The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 31:925–932
Heidinger-Pauli JM, Mert O, Davenport C, Guacci V, Koshland D (2010) Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr Biol 20:957–963
Hirota Y, Lahti JM (2000) Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase. Nucl Acids Res 28:917–924
Ivanov D, Schleiffer A, Eisenhaber F, Mechtler K, Haering CH, Nasmyth K (2002) Eco1 is a novel acetlytransferase that can acetylate proteins involved in cohesion. Curr Biol 12:323–328
Katou Y, Kanoh Y, Bandoh M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424:1078–1083
Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963–972
Laha S, Das SP, Hajra S, Sanyal K, Sinha P (2011) Functional characterization of the Saccharomyces cerevisiae protein Chl1 reveals the role of sister chromatid cohesion in the maintenance of spindle length during S-phase arrest. BMC Genet 12:83
Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430:573–578
Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner K-P, Shirahige K, Uhlmann F (2006) Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell 23:787–799
Lopez-Serra L, Lengronne A, Borges V, Kelly G, Uhlmann F (2013) Budding yeast Wapl controls sister chromatid cohesion maintenance and chromosome condensation. Curr Biol 23:64–69
Lyons NA, Morgan DO (2011) Cdk1-dependent destruction of Eco1 prevents cohesion establishment after S phase. Mol Cell 42:378–389
Mayer ML, Gygi SP, Aebersold R, Hieter P (2001) Identification of RFC(Ctf18p, Ctf8p, Dcc1p): An alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell 7:959–970
Mayer ML, Pot I, Chang M, Xu H, Aneliunas V, Kwok T, Newitt R, Aebersold R, Boone C, Brown GW, Hieter P (2004) Identification of protein complexes required for efficient sister chromatid cohesion. Mol Biol Cell 15:1736–1745
Michaelis C, Ciosk R, Nasmyth K (1997) Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35–45
Miles J, Formosa T (1992) Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase α, acts in DNA metabolism in vivo. Mol Cell Biol 12:5724–5735
Moldovan G-L, Pfander B, Jentsch S (2006) PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell 23:723–732
Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M (2009) An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6:917–922
Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, Peters J-M (2010) Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell 143:737–749
O'Reilly N, Charbin A, Lopez-Serra L, Uhlmann F (2012) Facile synthesis of budding yeast a-factor and its use to synchronize cells of α mating type. Yeast 29:233–240
Petronczki M, Chwalla B, Siomos MF, Yokobayashi S, Helmhart W, Deutschbauer AM, Davis RW, Watanabe Y, Nasmyth K (2004) Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-α-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J Cell Sci 117:3547–3559
Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouët F, Underwood P, Metson J, Imre R, Mechtler K, Katis VL, Nasmyth K (2009) Building sister chromatid cohesion: Smc3 acetylation counteracts an antiestablishment activity. Mol Cell 33:763–774
Rudra S, Skibbens RV (2012) Sister chromatid cohesion establishment occurs in concert with lagging strand synthesis. Cell Cycle 11:2114–2121
Skibbens RV (2004) Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion. Genetics 166:33–42
Skibbens RV, Corson LB, Koshland D, Hieter P (1999) Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev 13:307–319
Smith DJ, Whitehouse I (2012) Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 483:434–438
Strunnikov AV, Larionov VL, Koshland D (1993) SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J Cell Biol 123:1635–1648
Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K (2009) Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr Biol 19:492–497
Tanaka H, Katou Y, Yagura M, Saitoh K, Itoh T, Araki H, Bando M, Shirahige K (2009) Ctf4 coordinates the progression of helicase and DNA polymerase α. Genes Cells 14:807–820
Terret M-E, Sherwood R, Rahman S, Qin J, Jallepalli PV (2009) Cohesin acetylation speeds the replication fork. Nature 462:231–234
Thadani R, Uhlmann F, Heeger S (2012) Condensin, chromatin crossbarring and chromosome condensation. Curr Biol 22:R1012–R1021
Tóth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K (1999) Yeast Cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev 13:320–333
Uhlmann F (2009) A matter of choice: the establishment of sister chromatid cohesion. EMBO Rep 10:1095–1102
Uhlmann F, Lottspeich F, Nasmyth K (1999) Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400:37–42
Uhlmann F, Nasmyth K (1998) Cohesion between sister chromatids must be established during DNA replication. Curr Biol 8:1095–1101
Ünal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE (2008) A molecular determinant for the establishment of sister chromatid cohesion. Science 321:566–569
Wach A, Brachat A, Pöhlmann R, Philippsen P (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793–1808
Warren CD, Eckley EM, Lee MS, Hanna JS, Hughes A, Peyser B, Jie C, Irizarry R, Spencer FA (2004) S-phase checkpoint genes safeguard high-fidelity sister chromatid cohesion. Mol Biol Cell 15:1724–1735
Xu H, Boone C, Brown GW (2007) Genetic dissection of parallel sister-chromatid cohesion pathways. Genetics 176:1417–1429
Zhang J, Shi X, Li Y, Kim B-J, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, Zhang P, Kim S-T, Pan X, Qin J (2008) Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell 31:143–151
Zhou Y, Wang TS (2004) A coordinated temporal interplay of nucleosome reorganization factor, sister chromatin cohesion factor, and DNA polymerase α facilitates DNA replication. Mol Cell Biol 24:9568–9579
Acknowledgements
We are grateful to K. Shirahige for his gift of the ac-Smc3 antibody, J. Campbell for yeast strains and G. Kelly for biostatistics support. We thank the members of our laboratory for advice and comments on the manuscript. This work was funded by Cancer Research UK, the European Research Council (F.U., V. B.) and the National Institute of Health Grant R01 GM102253 to I.W. D.J.S. is a HHMI fellow of the Damon Runyon Cancer Research Foundation (DRG-#2046-10).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erich Nigg
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 679 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Borges, V., Smith, D.J., Whitehouse, I. et al. An Eco1-independent sister chromatid cohesion establishment pathway in S. cerevisiae . Chromosoma 122, 121–134 (2013). https://doi.org/10.1007/s00412-013-0396-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-013-0396-y